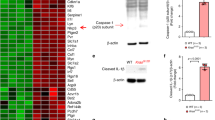
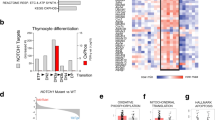
Abstract
The Notch signaling pathway contributes to the pathogenesis of a wide spectrum of human cancers, including hematopoietic malignancies. Its functions are highly dependent on the specific cellular context. Gain-of-function NOTCH1 mutations are prevalent in human T-cell leukemia, while loss of Notch signaling is reported in myeloid leukemias. Here, we report a novel oncogenic function of Notch signaling in oncogenic Kras-induced myeloproliferative neoplasm (MPN). We find that downregulation of Notch signaling in hematopoietic cells via DNMAML expression or Pofut1 deletion significantly blocks MPN development in KrasG12D mice in a cell-autonomous manner. Further mechanistic studies indicate that inhibition of Notch signaling upregulates Dusp1, a dual phosphatase that inactivates p-ERK, and downregulates cytokine-evoked ERK activation in KrasG12D cells. Moreover, mitochondrial metabolism is greatly enhanced in KrasG12D cells but significantly reprogrammed by DNMAML close to that in control cells. Consequently, cell proliferation and expanded myeloid compartment in KrasG12D mice are significantly reduced. Consistent with these findings, combined inhibition of the MEK/ERK pathway and mitochondrial oxidative phosphorylation effectively inhibited the growth of human and mouse leukemia cells in vitro. Our study provides a strong rational to target both ERK signaling and aberrant metabolism in oncogenic Ras-driven myeloid leukemia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–81.
Kidd S, Kelley MR, Young MW. Sequence of the notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factors. Mol Cell Biol. 1986;6:3094–108.
Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell. 2014;25:318–34.
Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–75.
Wang Y, Shao L, Shi S, Harris RJ, Spellman MW, Stanley P, et al. Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J Biol Chem. 2001;276:40338–45.
Luo Y, Haltiwanger RS. O-fucosylation of notch occurs in the endoplasmic reticulum. J Biol Chem. 2005;280:11289–94.
Okajima T, Xu A, Lei L, Irvine KD. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science. 2005;307:1599–603.
Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002;111:893–904.
Maillard I, Koch U, Dumortier A, Shestova O, Xu L, Sai H, et al. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2008;2:356–66.
Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6.
Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71.
Kong G, Du J, Liu Y, Meline B, Chang YI, Ranheim EA, et al. Notch1 gene mutations target KRAS G12D-expressing CD8+ cells and contribute to their leukemogenic transformation. J Biol Chem. 2013;288:18219–27.
Ashworth TD, Pear WS, Chiang MY, Blacklow SC, Mastio J, Xu L, et al. Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1. Blood. 2010;116:5455–64.
Chiang MY, Xu L, Shestova O, Histen G, L’Heureux S, Romany C, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118:3181–94.
Kannan S, Sutphin RM, Hall MG, Golfman LS, Fang W, Nolo RM, et al. Notch activation inhibits AML growth and survival: a potential therapeutic approach. J Exp Med. 2013;210:321–37.
Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N, et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J Exp Med. 2013;210:301–19.
Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–3.
Yao D, Huang Y, Huang X, Wang W, Yan Q, Wei L, et al. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood. 2011;117:5652–62.
Zhang J, Wang J, Liu Y, Sidik H, Young KH, Lodish HF, et al. Oncogenic Kras-induced leukemogeneis: hematopoietic stem cells as the initial target and lineage-specific progenitors as the potential targets for final leukemic transformation. Blood. 2009;113:1304–14.
Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci USA. 2003;100:5234–9.
Du J, Liu Y, Meline B, Kong G, Tan LX, Lo JC, et al. Loss of CD44 attenuates aberrant GM-CSF signaling in Kras G12D hematopoietic progenitor/precursor cells and prolongs the survival of diseased animals. Leukemia. 2013;27:754–7.
Chang YI, You X, Kong G, Ranheim EA, Wang J, Du J, et al. Loss of Dnmt3a and endogenous Kras cooperate to regulate hematopoietic stem and progenitor cell functions in leukemogenesis. Leukemia. 2015;29:1847–56.
Kong G, Chang Y-I, Damnernsawad A, You X, Du J, Ranheim RA, et al. Loss of wild-type Kras promotes activation of all Ras isoforms in oncogenic Kras-induced leukemogenesis. Leukemia. 2016;30:1542–51.
Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–77.
Ye M, Iwasaki H, Laiosa CV, Stadtfeld M, Xie H, Heck S, et al. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity. 2003;19:689–99.
Oh P, Lobry C, Gao J, Tikhonova A, Loizou E, Manent J, et al. In vivo mapping of notch pathway activity in normal and stress hematopoiesis. Cell Stem Cell. 2013;13:190–204.
Van Meter ME, Diaz-Flores E, Archard JA, Passegue E, Irish JM, Kotecha N, et al. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood. 2007;109:3945–52.
Maraver A, Fernandez-Marcos PJ, Herranz D, Canamero M, Munoz-Martin M, Gomez-Lopez G, et al. Therapeutic effect of gamma-secretase inhibition in KrasG12V-driven non-small cell lung carcinoma by derepression of DUSP1 and inhibition of ERK. Cancer Cell. 2012;22:222–34.
Zhang J, Kong G, Rajagopalan A, Lu L, Song J, Hussaini M, et al. p53-/- synergizes with enhanced NrasG12D signaling to transform megakaryocyte-erythroid progenitors in acute myeloid leukemia. Blood. 2017;129:358–70.
Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261–6.
Grieselhuber NR, Klco JM, Verdoni AM, Lamprecht T, Sarkaria SM, Wartman LD, et al. Notch signaling in acute promyelocytic leukemia. Leukemia. 2013;27:1548–57.
Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci USA. 2011;108:14908–13.
Herranz D, Ambesi-Impiombato A, Sudderth J, Sanchez-Martin M, Belver L, Tosello V, et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med. 2015;21:1182–9.
Yeh TC, Marsh V, Bernat BA, Ballard J, Colwell H, Evans RJ, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res. 2007;13:1576–83.
Kong G, Wunderlich M, Yang D, Ranheim EA, Young KH, Wang J, et al. Combined MEK and JAK inhibition abrogates murine myeloproliferative neoplasm. J Clin Invest. 2014;124:2762–73.
Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell. 2011;44:864–77.
Kindler T, Cornejo MG, Scholl C, Liu J, Leeman DS, Haydu JE, et al. K-RasG12D-induced T-cell lymphoblastic lymphoma/leukemias harbor Notch1 mutations and are sensitive to gamma-secretase inhibitors. Blood. 2008;112:3373–82.
Chang YI, Damnernsawad A, Allen LK, Yang D, Ranheim EA, Young KH, et al. Evaluation of allelic strength of human TET2 mutations and cooperation between Tet2 knockdown and oncogenic Nras mutation. Br J Haematol. 2014;166:461–5.
Merlevede J, Droin N, Qin T, Meldi K, Yoshida K, Morabito M, et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat Commun. 2016;7:10767.
Ambrogio C, Gomez-Lopez G, Falcone M, Vidal A, Nadal E, Crosetto N, et al. Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma. Nat Med. 2016;22:270–7.
Xu J, Chi F, Guo T, Punj V, Lee WN, French SW, et al. NOTCH reprograms mitochondrial metabolism for proinflammatory macrophage activation. J Clin Invest. 2015;125:1579–90.
Kishton RJ, Barnes CE, Nichols AG, Cohen S, Gerriets VA, Siska PJ, et al. AMPK is essential to balance glycolysis and mitochondrial metabolism to control T-ALL cell stress and survival. Cell Metab. 2016;23:649–62.
Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–41.
Jacque N, Ronchetti AM, Larrue C, Meunier G, Birsen R, Willems L, et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood. 2015;126:1346–56.
Liu Y, Marks K, Cowley GS, Carretero J, Liu Q, Nieland TJ, et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer Discov. 2013;3:870–9.
Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531:110–3.
Castro-Malaspina H, Schaison G, Passe S, Pasquier A, Berger R, Bayle-Weisgerber C, et al. Subacute and chronic myelomonocytic leukemia in children (juvenile CML). Clinical and hematologic observations, and identification of prognostic factors. Cancer. 1984;54:675–86.
Passmore SJ, Hann IM, Stiller CA, Ramani P, Swansbury GJ, Gibbons B, et al. Pediatric myelodysplasia: a study of 68 children and a new prognostic scoring system. Blood. 1995;85:1742–50.
Passmore SJ, Chessells JM, Kempski H, Hann IM, Brownbill PA, Stiller CA. Paediatric myelodysplastic syndromes and juvenile myelomonocytic leukaemia in the UK: a population-based study of incidence and survival. Br J Haematol. 2003;121:758–67.
Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–81.
Acknowledgements
We are grateful to Patrick Nyman and Dr. Paul Lambert for providing the Rosa26LSL DNMAML-GFP/+ mice and to Dr. Pamela Stanley for sharing the Pofut1fl/fl mice. We appreciate the critical comments from Drs. Emery Bresnick and Paul Lambert on the manuscript. We would like to thank the University of Wisconsin Carbone Comprehensive Cancer Center (UWCCC) for use of its Shared Services (Flow Cytometry Laboratory and Experimental Pathology Laboratory) to complete this research. This work was supported by the National Natural Science Foundation of China (NO.81600100) to G.K., Alexander von Humboldt Foundation (Alfred Toepfer Faculty Fellow) and NIH-MIRA grant R35GM124806 to X.Z., grants from American Cancer Society LIB-125064 and NIH HL103827 to L.Z., and R01 grants CA152108 and HL113066, and a Scholar Award from the Leukemia & Lymphoma Society to J.Z.. This work was also supported in part by NIH/NCI P30 CA014520--UW Comprehensive Cancer Center Support.
Author contributions
Conception and design: G. Kong, X. You, and J. Zhang. Acquisition of data: G. Kong, X. You, Z. Wen, Y.-I Chang, C. Letson, J. F. Zhang, Y. Zhou, Y. Liu, A. Rajagopalan. Analysis and interpretation of data: G. Kong, J. F. Zhang, X. Zhang, E. A. Ranheim, A. Rajagopalan, E. Padron, W. S. Pear, L. Zhou, and J. Zhang. Writing, review, and/or revision of the manuscript: G. Kong, E. A. Ranheim, E. Stieglitz, M. Loh, W. S. Pear, L. Zhou, and J. Zhang. Technical or material support: S. Qian, I. Hofmann-Zhang, D. Yang, E. Stieglitz, M. Loh, X. Zhong, W. S. Pear and L. Zhou. Study supervision: J. Zhang
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Kong, G., You, X., Wen, Z. et al. Downregulating Notch counteracts KrasG12D-induced ERK activation and oxidative phosphorylation in myeloproliferative neoplasm. Leukemia 33, 671–685 (2019). https://doi.org/10.1038/s41375-018-0248-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-018-0248-0