Abstract
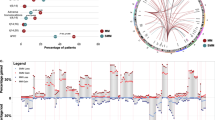
Clonal evolution drives tumor progression, chemoresistance and relapse in cancer. Little is known about clonal selection induced by therapeutic pressure in multiple myeloma. To address this issue, we performed large targeted sequencing of bone marrow plasma cells in 43 multiple myeloma patients at diagnosis and at relapse from exactly the same intensive treatment. The most frequently mutated genes at diagnosis were KRAS (35%), NRAS (28%), DIS3 (16%), BRAF, and LRP1B (12% each). At relapse, the mutational burden was unchanged. Many of the mutations were present at the subclonal level at both time points, including driver ones. According to patients and mutations, we observed different scenarios: selection of a very rare subclone present at diagnosis, appearance, or disappearance of mutations, but also stability. Our data highlight that chemoresistance and relapse could be induced by newly acquired mutations in myeloma drivers but also by (sub)clonal mutations preexisting to the treatment. Importantly, no specific mutation or rearrangement was observed at relapse, demonstrating that intensive treatment has a nonspecific effect on clonal selection in multiple myeloma. Finally, we identified 22 cases of biallelic event, including a double event deletion 17p/TP53mut.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–60.
Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997.
Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101.
Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33:3911–20.
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21.
Egan JB, Shi C-X, Tembe W, Christoforides A, Kurdoglu A, Sinari S, et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood 2012. 2012;120:1060–6.
Walker BA, Wardell CP, Melchor L, Hulkki S, Potter NE, Johnson DC, et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012;120:1077–86.
Keats JJ, Chesi M, Egan JB, Garbitt VM, Palmer SE, Braggio E, et al. Clonal competition with alternating dominance in multiple myeloma. Blood 2012. 2012;120:1067–76.
Magrangeas F, Avet-Loiseau H, Gouraud W, Lode L, Decaux O, Godmer P, et al. Minor clone provides a reservoir for relapse in multiple myeloma. Leukemia. 2013;27:473–81.
Walker BA, Wardell CP, Melchor L, Brioli A, Johnson DC, Kaiser MF, et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28:384–90.
Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–48.
Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8.
Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–45.
Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13.
Corre J, Munshi N, Avet-Loiseau H. Genetics of multiple myeloma: another heterogeneity level? Blood. 2015;125:1870–6.
Robiou du Pont S, Cleynen A, Fontan C, Attal M, Munshi N, Corre J, et al. Genomics of multiple myeloma. J Clin Oncol. 2017;35:963–7.
Bolli N, Li Y, Sathiaseelan V, Raine K, Jones D, Ganly P, et al. A DNA target-enrichment approach to detect mutations, copy number changes and immunoglobulin translocations in multiple myeloma. Blood Cancer J. 2016;6:e467.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–95.
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucl Acids Res. 2010;38:e164.
Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–81.
Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–10.
Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–93.
Devarakonda S, Govindan R. Clonal evolution: multiregion sequencing of esophageal adenocarcinoma before and after chemotherapy. Cancer Discov. 2015;5:796–8.
Weinhold N, Ashby C, Rasche L, Chavan SS, Stein C, Stephens OW, et al. Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood. 2016;128:1735–44.
McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26.
O’Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood. 2007;110:2242–9.
Woyach JA, Furman RR, Liu TM, Ozer HG, Zapatka M, Ruppert AS, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370:2286–94.
Amirouchene-Angelozzi N, Swanton C, Bardelli A. Tumor evolution as a therapeutic target. Cancer Discover. 2017. Cancer Discovery 2017;7:805-17.
Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21.
Kortum KM, Langer C, Monge J, Bruins L, Egan JB, Zhu YX, et al. Targeted sequencing using a 47 gene multiple myeloma mutation panel (M(3) P) in -17p high risk disease. Br J Haematol. 2015;168:507–10.
Kortum KM, Mai EK, Hanafiah NH, Shi CX, Zhu YX, Bruins L, et al. Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood. 2016;128:1226–33.
Surget S, Lemieux-Blanchard E, Maiga S, Descamps G, Le Gouill S, Moreau P, et al. Bendamustine and melphalan kill myeloma cells similarly through reactive oxygen species production and activation of the p53 pathway and do not overcome resistance to each other. Leuk Lymphoma. 2014;55:2165–73.
Lu S, Wang J. The resistance mechanisms of proteasome inhibitor bortezomib. Biomark Res. 2013;1:13.
Keats J, Fonseca R, Chesi M, Schop R, Baker A, Chng W, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–44.
Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–7.
de Haart SJ, Willems SM, Mutis T, Koudijs MJ, van Blokland MT, Lokhorst HM, et al. Comparison of intramedullary myeloma and corresponding extramedullary soft tissue plasmacytomas using genetic mutational panel analyses. Blood Cancer J. 2016;6:e426.
Rasche L, Chavan SS, Stephens OW, Patel PH, Tytarenko R, Ashby C, et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat Commun. 2017;8:268.
Acknowledgements
This work was supported by NIH grants PO1-155258 and P50-100707 (NM, HAL); Department of Veterans Affairs Merit Review Award 1 I01BX001584-01 (NM) and the CAPTOR program. The CRCT Team 13 is labeled by ARC. We thank the Intergroupe Francophone du Myelome for providing patient samples and clinical data.
Author contributions
JC, AC, NM, and HAL conceived the project, JC, AC, SR, and HAL analyzed the data, AC performed bioinformatics analysis, LB performed experiments, NB conceived the sequencing targeted panel, MA provided samples and clinical data, AC, JC, NM, and HAL wrote the manuscript which was reviewed and edited by the other co-authors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
NB received honoraria and personal fees from Celgene Corporation. The remaining authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Corre, J., Cleynen, A., Robiou du Pont, S. et al. Multiple myeloma clonal evolution in homogeneously treated patients. Leukemia 32, 2636–2647 (2018). https://doi.org/10.1038/s41375-018-0153-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-018-0153-6
This article is cited by
-
Fluorescence in situ hybridization reveals the evolutionary biology of minor clone of gain/amp(1q) in multiple myeloma
Leukemia (2024)
-
Dynamic single-cell RNA-seq analysis reveals distinct tumor program associated with microenvironmental remodeling and drug sensitivity in multiple myeloma
Cell & Bioscience (2023)
-
Mass Cytometry reveals unique phenotypic patterns associated with subclonal diversity and outcomes in multiple myeloma
Blood Cancer Journal (2023)
-
UTX inactivation in germinal center B cells promotes the development of multiple myeloma with extramedullary disease
Leukemia (2023)
-
Clonal evolution after treatment pressure in multiple myeloma: heterogenous genomic aberrations and transcriptomic convergence
Leukemia (2022)