Abstract
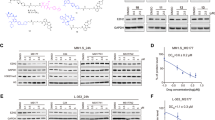
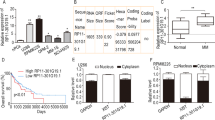
EZH2 is highly expressed in multiple myeloma (MM). However, the molecular mechanisms underlying EZH2 overexpression and its role in drug resistance of MM remain undefined. Here we show that EZH2 is upregulated in drug-resistant MM cells and its aberrant overexpression is associated with poor prognosis of MM patients. Overexpression of EZH2 in parental MM cells renders them resistant to anti-myeloma drugs and suppression of EZH2 displays the opposite effects. Using miRNA target scan algorithms, we identify miR-138 as a regulator of EZH2, which is conversely repressed by EZH2-induced H3K27 trimethylation in MM-resistant cell lines and primary tumor cells. Analysis of ChIP-seq dataset and H3K27me3 ChIP reveals that RBPMS is a direct and functionally relevant target of EZH2. RBPMS silencing confers resistance to MM cells and restoration of RBPMS by miR-138 overexpression re-sensitizes the resistant cells to drug. Importantly, in vivo delivery of miR-138 mimics or pharmacological inhibitor of EZH2 in combination with a proteasome inhibitor, bortezomib, induces significant regression of tumors in xenograft model. This study establishes EZH2/miR-138 axis as a potential therapeutic target for MM.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Dimopoulos K, Gimsing P, Gronbaek K. The role of epigenetics in the biology of multiple myeloma. Blood Cancer J. 2014;4:e207.
Merin NM, Kelly KR. Clinical use of proteasome inhibitors in the treatment of multiple myeloma. Pharm (Basel). 2014;8:1–20.
Abdi J, Chen G, Chang H. Drug resistance in multiple myeloma: latest findings and new concepts on molecular mechanisms. Oncotarget. 2013;4:2186–207.
Pawlyn CKM, Stein CK, Wardell ChP, Macleod V, Edmondson R, Barlogie B, et al. EZH2 overexpression in myeloma patients shortens survival and in-vitro data supports a potential new targeted treatment strategy. Cancer Res. 2015;75:3515.
Volkel P, Dupret B, Le Bourhis X, Angrand PO. Diverse involvement of EZH2 in cancer epigenetics. Am J Transl Res. 2015;7:175–93.
Lund K, Adams PD, Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia. 2014;28:44–49.
Beguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23:677–92.
Pawlyn C, Bright MD, Buros AF, Stein CK, Walters Z, Aronson LI, et al. Overexpression of EZH2 in multiple myeloma is associated with poor prognosis and dysregulation of cell cycle control. Blood Cancer J. 2017;7:e549.
Agarwal P, Alzrigat M, Parraga AA, Enroth S, Singh U, Ungerstedt J, et al. Genome-wide profiling of histone H3 lysine 27 and lysine 4 trimethylation in multiple myeloma reveals the importance of Polycomb gene targeting and highlights EZH2 as a potential therapeutic target. Oncotarget. 2016;7:6809–23.
Kalushkova A, Fryknas M, Lemaire M, Fristedt C, Agarwal P, Eriksson M, et al. Polycomb target genes are silenced in multiple myeloma. PLoS One. 2010;5:e11483.
Zeng D, Liu M, Pan J. Blocking EZH2 methylation transferase activity by GSK126 decreases stem cell-like myeloma cells. Oncotarget. 2017;8:3396–411.
Hernando H, Gelato KA, Lesche R, Beckmann G, Koehr S, Otto S, et al. EZH2 inhibition blocks multiple myeloma cell growth through upregulation of epithelial tumor suppressor genes. Mol Cancer Ther. 2016;15:287–98.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–8.
Barbarotto E, Calin GA. Potential therapeutic applications of miRNA-based technology in hematological malignancies. Curr Pharm Des. 2008;14:2040–50.
Mills K. Gene expression profiling for the diagnosis and prognosis of acute myeloid leukaemia. Front Biosci. 2008;13:4605–16.
Ritchie WJ, Flamant S, Rasko JE. MicroRNA in acute myeloid leukemia. N Engl J Med. 2008;359:653–4.
Calin GA, Croce CM. Investigation of microRNA alterations in leukemias and lymphomas. Methods Enzymol. 2007;427:193–213.
Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA. 2008;105:12885–90.
Duan W, Gao L, Wu X, Wang L, Nana-Sinkam SP, Otterson GA, et al. MicroRNA-34a is an important component of PRIMA-1-induced apoptotic network in human lung cancer cells. Int J Cancer. 2010;127:313–20.
Misiewicz-Krzeminska I, Sarasquete ME, Quwaider D, Krzeminski P, Ticona FV, Paino T, et al. Restoration of microRNA-214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica. 2013;98:640–8.
Zhang YK, Wang H, Leng Y, Li ZL, Yang YF, Xiao FJ, et al. Overexpression of microRNA-29b induces apoptosis of multiple myeloma cells through down regulating Mcl-1. Biochem Biophys Res Commun. 2011;414:233–9.
Shi M, Guo N. MicroRNA expression and its implications for the diagnosis and therapeutic strategies of breast cancer. Cancer Treat Rev. 2009;35:328–34.
Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63.
Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69.
Buzzeo R, Enkemann S, Nimmanapalli R, Alsina M, Lichtenheld MG, Dalton WS, et al. Characterization of a R115777-resistant human multiple myeloma cell line with cross-resistance to PS-341. Clin Cancer Res. 2005;11:6057–64.
Yang Y, Chen Y, Saha MN, Chen J, Evans K, Qiu L, et al. Targeting phospho-MARCKS overcomes drug-resistance and induces antitumor activity in preclinical models of multiple myeloma. Leukemia. 2015;29:715–26.
Saha MN, Chen Y, Chen MH, Chen G, Chang H. Small molecule MIRA-1 induces in vitro and in vivo anti-myeloma activity and synergizes with current anti-myeloma agents. Br J Cancer. 2014;110:2224–31.
Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–7.
Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–32.
Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–153.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Chang JW, Gwak SY, Shim GA, Liu L, Lim YC, Kim JM, et al. EZH2 is associated with poor prognosis in head-and-neck squamous cell carcinoma via regulating the epithelial-to-mesenchymal transition and chemosensitivity. Oral Oncol. 2016;52:66–74.
Wassef M, Michaud A, Margueron R. Association between EZH2 expression, silencing of tumor suppressors and disease outcome in solid tumors. Cell Cycle. 2016;15:2256–62.
Croonquist PA, Van Ness B. The polycomb group protein enhancer of zeste homolog 2 (EZH 2) is an oncogene that influences myeloma cell growth and the mutant ras phenotype. Oncogene. 2005;24:6269–80.
Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–9.
Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9.
Wang L, Zhang X, Jia LT, Hu SJ, Zhao J, Yang JD, et al. c-Myc-mediated epigenetic silencing of MicroRNA-101 contributes to dysregulation of multiple pathways in hepatocellular carcinoma. Hepatology. 2014;59:1850–63.
Zhuang C, Wang P, Huang D, Xu L, Wang X, Wang L, et al. A double-negative feedback loop between EZH2 and miR-26a regulates tumor cell growth in hepatocellular carcinoma. Int J Oncol. 2016;48:1195–204.
Alzrigat M, Parraga AA, Agarwal P, Zureigat H, Osterborg A, Nahi H, et al. EZH2 inhibition in multiple myeloma downregulates myeloma associated oncogenes and upregulates microRNAs with potential tumor suppressor functions. Oncotarget. 2017;8:10213–24.
Cai S, Zhou P, Liu Z. Functional characteristics of a double negative feedback loop mediated by microRNAs. Cogn Neurodyn. 2013;7:417–29.
Báez-Vega PM, Echevarría-Vargas IM, Valiyeva F, Encarnación-Rosado J, Roman A, Flores J, et al. Targeting miR-21-3p inhibits proliferation and invasion of ovarian cancer cells. Oncotarget. 2016;7:36321–37.
Baughn LB, Di Liberto M, Niesvizky R, Cho HJ, Jayabalan D, Lane J, et al. CDK2 phosphorylation of Smad2 disrupts TGF-beta transcriptional regulation in resistant primary bone marrow myeloma cells. J Immunol. 2009;182:1810–7.
Sun Y, Ding L, Zhang H, Han J, Yang X, Yan J, et al. Potentiation of Smad-mediated transcriptional activation by the RNA-binding protein RBPMS. Nucleic Acids Res. 2006;34:6314–26.
Ahmad N, Haider S, Jagannathan S, Anaissie E, Driscoll JJ. MicroRNA theragnostics for the clinical management of multiple myeloma. Leukemia. 2014;28:732–8.
Tagliaferri P, Rossi M, Di Martino MT, Amodio N, Leone E, Gulla A, et al. Promises and challenges of MicroRNA-based treatment of multiple myeloma. Curr Cancer Drug Targets. 2012;12:838–46.
Nana-Sinkam SP, Croce CM. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther. 2013;93:98–104.
Di Martino MT, Leone E, Amodio N, Foresta U, Lionetti M, Pitari MR, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res. 2012;18:6260–70.
Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19:1116–22.
Rizq O, Mimura N, Oshima M, Saraya A, Koide S, Kato Y, et al. Dual inhibition of EZH2 and EZH1 sensitizes PRC2-dependent tumors to proteasome inhibition. Clin Cancer Res. 2017;23:4817–30.
Acknowledgements
The study was supported in part by the grants from Cancer Research Society (CRS) and Leukemia & Lymphoma Society of Canada (LLSC).
Author contributions
HC and NR conceived and designed the study. NR, MP, and JA carried out experiments and statistical analysis. DR provided MM patient clinical information. NR and HC wrote the manuscript. HC supervised the project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Rastgoo, N., Pourabdollah, M., Abdi, J. et al. Dysregulation of EZH2/miR-138 axis contributes to drug resistance in multiple myeloma by downregulating RBPMS. Leukemia 32, 2471–2482 (2018). https://doi.org/10.1038/s41375-018-0140-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-018-0140-y
This article is cited by
-
Melatonin and 5-fluorouracil combination chemotherapy: opportunities and efficacy in cancer therapy
Cell Communication and Signaling (2023)
-
Tumor-suppressive functions of protein lysine methyltransferases
Experimental & Molecular Medicine (2023)
-
Progranulin regulates the development and function of NKT2 cells through EZH2 and PLZF
Cell Death & Differentiation (2022)
-
Methylation-dependent and -independent roles of EZH2 synergize in CDCA8 activation in prostate cancer
Oncogene (2022)
-
EZH2 is associated with cartilage degeneration in osteoarthritis by promoting SDC1 expression via histone methylation of the microRNA-138 promoter
Laboratory Investigation (2021)