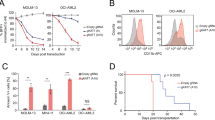
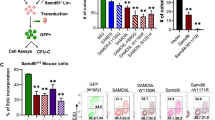
Abstract
ASXL1 plays key roles in epigenetic regulation of gene expression through methylation of histone H3K27, and disruption of ASXL1 drives myeloid malignancies, at least in part, via derepression of posterior HOXA loci. However, little is known about the identity of proteins that interact with ASXL1 and about the functions of ASXL1 in modulation of the active histone mark, such as H3K4 methylation. In this study, we demonstrate that ASXL1 is a part of a protein complex containing HCFC1 and OGT; OGT directly stabilizes ASXL1 by O-GlcNAcylation. Disruption of this novel axis inhibited myeloid differentiation and H3K4 methylation as well as H2B glycosylation and impaired transcription of genes involved in myeloid differentiation, splicing, and ribosomal functions; this has implications for myelodysplastic syndrome (MDS) pathogenesis, as each of these processes are perturbed in the disease. This axis is responsible for tumor suppression in the myeloid compartment, as reactivation of OGT induced myeloid differentiation and reduced leukemogenecity both in vivo and in vitro. Our data also suggest that MLL5, a known HCFC1/OGT-interacting protein, is responsible for gene activation by the ASXL1–OGT axis. These data shed light on the novel roles of the ASXL1–OGT axis in H3K4 methylation and activation of transcription.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Fisher CL, Pineault N, Brookes C, Helgason CD, Ohta H, Bodner C, et al. Loss-of-function Additional sex combs like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia. Blood. 2010;115:38–46.
Sinclair DA, Milne TA, Hodgson JW, Shellard J, Salinas CA, Kyba M, et al. The Additional sex combs gene of Drosophila encodes a chromatin protein that binds to shared and unique Polycomb group sites on polytene chromosomes. Development. 1998;125:1207–16.
Baskind HA, Na L, Ma Q, Patel MP, Geenen DL, Wang QT. Functional conservation of Asxl2, a murine homolog for the Drosophila enhancer of trithorax and polycomb group gene Asx. PLOS One. 2009;4:e4750.
Fisher CL, Randazzo F, Humphries RK, Brock HW. Characterization of Asxl1, a murine homolog of Additional sex combs, and analysis of the Asx-like gene family. Gene. 2006;369:109–18.
Fisher CL, Berger J, Randazzo F, Brock HW. A human homolog of Additional sex combs, Additional sex combs-like 1, maps to chromosome 20q11. Gene. 2003;306:115–26.
Abdel-Wahab O, Patel J, Levine RL. Clinical implications of novel mutations in epigenetic modifiers in AML. Hematol Oncol Clin North Am. 2011;25:1119–33.
Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol: Off J Am Soc Clin Oncol. 2011;29:504–15.
Gelsi-Boyer V, Trouplin V, Adelaide J, Bonansea J, Cervera N, Carbuccia N, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145:788–800.
Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69.
Gelsi-Boyer V, Brecqueville M, Devillier R, Murati A, Mozziconacci MJ, Birnbaum D. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol Oncol. 2012;5:12.
Thol F, Friesen I, Damm F, Yun H, Weissinger EM, Krauter J, et al. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J Clin Oncol. 2011;29:2499–506.
Inoue D, Kitaura J, Togami K, Nishimura K, Enomoto Y, Uchida T, et al. Myelodysplastic syndromes are induced by histone methylation-altering ASXL1 mutations. J Clin Invest. 2013;123:4627–40.
Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210:2641–59.
Bejar R, Lord A, Stevenson K, Bar-Natan M, Perez-Ladaga A, Zaneveld J, et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124:2705–12.
Wang JP, Li ZM, He YZ, Pan F, Chen S, Rhodes S, et al. Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood. 2014;123:541–53.
Balasubramani A, Larjo A, Bassein JA, Chang X, Hastie RB, Togher SM, et al. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat Commun. 2015;6:7307.
Shi H, Yamamoto S, Sheng M, Bai J, Zhang P, Chen R, et al. ASXL1 plays an important role in erythropoiesis. Sci Rep. 2016;6:28789.
Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, et al. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci USA. 2014;111:1355–60.
Zhou P, Wang Z, Yuan X, Zhou C, Liu L, Wan X, et al. Mixed lineage leukemia 5 (MLL5) protein regulates cell cycle progression and E2F1-responsive gene expression via association with host cell factor-1 (HCF-1). J Biol Chem. 2013;288:17532–43.
Lewis BA, Hanover JA. O-GlcNAc and the epigenetic regulation of gene expression. J Biol Chem. 2014;289:34440–8.
Ma Z, Vosseller K. Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J Biol Chem. 2014;289:34457–65.
Ma Z, Vocadlo DJ, Vosseller K. Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-kappaB activity in pancreatic cancer cells. J Biol Chem. 2013;288:15121–30.
Itkonen HM, Minner S, Guldvik IJ, Sandmann MJ, Tsourlakis MC, Berge V, et al. O-GlcNAc transferase integrates metabolic pathways to regulate the stability of c-MYC in human prostate cancer cells. Cancer Res. 2013;73:5277–87.
Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer. 2011;11:678–84.
Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct Funct. 2008;33:61–73.
Inoue D, Nishimura K, Kozuka-Hata H, Oyama M, Kitamura T. The stability of epigenetic factor ASXL1 is regulated through ubiquitination and USP7-mediated deubiquitination. Leukemia. 2015;29:2257–60.
Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–6.
Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–93.
Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–55.
Ding XD, Jiang W, Zhou PP, Liu LL, Wan XL, Yuan XJ, et al. Mixed Lineage Leukemia 5 (MLL5) protein stability is cooperatively regulated by O-GlcNac transferase (OGT) and ubiquitin specific protease 7 (USP7). PLOS One 2015;10:e0145023.
Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49:645–56.
Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–4.
Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Bi Y, Gong M, He Y, Zhang X, Zhou X, Zhang Y, et al. AP2alpha transcriptional activity is essential for retinoid-induced neuronal differentiation of mesenchymal stem cells. Int J Biochem Cell Biol. 2014;46:148–60.
Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–48.
Rosmarin AG, Luo M, Caprio DG, Shang J, Simkevich CP. Sp1 cooperates with the ets transcription factor, GABP, to activate the CD18 (beta2 leukocyte integrin) promoter. J Biol Chem. 1998;273:13097–103.
Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T. MafB is an inducer of monocytic differentiation. EMBO J. 2000;19:1987–97.
Gibbs JD, Liebermann DA, Hoffman B. Egr-1 abrogates the E2F-1 block in terminal myeloid differentiation and suppresses leukemia. Oncogene. 2008;27:98–106.
Yang ZF, Drumea K, Cormier J, Wang J, Zhu X, Rosmarin AG. GABP transcription factor is required for myeloid differentiation, in part, through its control of Gfi-1 expression. Blood. 2011;118:2243–53.
Daou S, Hammond-Martel I, Mashtalir N, Barbour H, Gagnon J, Iannantuono NV, et al. The BAP1/ASXL2 histone H2A deubiquitinase complex regulates cell proliferation and is disrupted in cancer. J Biol Chem. 2015;290:28643–63.
Vogel JL, Kristie TM. The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J. 2000;19:683–90.
Chen HM, Pahl HL, Scheibe RJ, Zhang DE, Tenen DG. The Sp1 transcription factor binds the CD11b promoter specifically in myeloid cells in vivo and is essential for myeloid-specific promoter activity. J Biol Chem. 1993;268:8230–9.
Shi FT, Kim H, Lu W, He Q, Liu D, Goodell MA, et al. Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J Biol Chem. 2013;288:20776–84.
Zachara NE, Hart GW. O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim Biophys Acta. 2004;1673:13–28.
Damm F, Oberacker T, Thol F, Surdziel E, Wagner K, Chaturvedi A, et al. Prognostic importance of histone methyltransferase MLL5 expression in acute myeloid leukemia. J Clin Oncol. 2011;29:682–9.
Sashida G, Harada H, Matsui H, Oshima M, Yui M, Harada Y, et al. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat Commun. 2014;5:4177.
Kondo Y. Targeting histone methyltransferase EZH2 as cancer treatment. J Biochem. 2014;156:249–57.
Ruggero D, Shimamura A. Marrow failure: a window into ribosome biology. Blood. 2014;124:2784–92.
Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, et al. Identification of RPS14 as a 5q-syndrome gene by RNA interference screen. Nature. 2008;451:335–9.
Cai X, Gao L, Teng L, Ge J, Oo ZM, Kumar AR. et al. Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell. 2015;17:165–77.
Obeng EA, Chappell RJ, Seiler M, Chen MC, Campagna DR, Schmidt PJ, et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016;30:404–17.
Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SCW, Ramakrishnan A, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27:617–30.
Shirai CL, Ley JN, White BS, Kim S, Tibbitts J, Shao J, et al. Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell. 2015;27:631–43.
Acknowledgements
We thank LW. Deng and X. Yu for providing plasmids. This work is supported by the Grants-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, A research grant from the Tokyo Biochemical Research Foundation, and A research grant from the Uehara Memorial Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Inoue, D., Fujino, T., Sheridan, P. et al. A novel ASXL1–OGT axis plays roles in H3K4 methylation and tumor suppression in myeloid malignancies. Leukemia 32, 1327–1337 (2018). https://doi.org/10.1038/s41375-018-0083-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-018-0083-3
This article is cited by
-
O-GlcNAcylation: the sweet side of epigenetics
Epigenetics & Chromatin (2023)
-
Epigenetic regulation by ASXL1 in myeloid malignancies
International Journal of Hematology (2023)
-
ASXL1/2 mutations and myeloid malignancies
Journal of Hematology & Oncology (2022)
-
O-GlcNAcylation: an important post-translational modification and a potential therapeutic target for cancer therapy
Molecular Medicine (2022)
-
Molecular Pathogenesis of Myeloproliferative Neoplasms
Current Hematologic Malignancy Reports (2022)