Abstract
Gastric cancer possesses great histological and molecular diversity, which creates obstacles for rapid and efficient diagnoses. Classic diagnoses either depend on the pathologist’s judgment, which relies heavily on subjective experience, or time-consuming molecular assays for subtype diagnosis. Here, we present a deep learning (DL) system to achieve interpretable tumor differentiation grade and microsatellite instability (MSI) recognition in gastric cancer directly using hematoxylin-eosin (HE) staining whole-slide images (WSIs). WSIs from 467 patients were divided into three cohorts: the training cohort with 348 annotated WSIs, the testing cohort with 88 annotated WSIs, and the integration testing cohort with 31 original WSIs without tumor contour annotation. First, the DL models comprehensibly achieved tumor differentiation recognition with an F1 values of 0.8615 and 0.8977 for poorly differentiated adenocarcinoma (PDA) and well-differentiated adenocarcinoma (WDA) classes. Its ability to extract pathological features about the glandular structure formation, which is the key to distinguishing between PDA and WDA, increased the interpretability of the DL models. Second, the DL models achieved MSI status recognition with a patient-level accuracy of 86.36% directly from HE-stained WSIs in the testing cohort. Finally, the integrated end-to-end system achieved patient-level MSI recognition from original HE staining WSIs with an accuracy of 83.87% in the integration testing cohort with no tumor contour annotation. The proposed system, therefore, demonstrated high accuracy and interpretability, which can potentially promote the implementation of artificial intelligence healthcare.
Similar content being viewed by others
Introduction
Deep learning (DL) technologies have been increasingly studied in the healthcare field1,2, especially in cancer diagnosis3,4,5. Molecular changes in cancer tissues often lead to changes in the morphology of tumor cells and their microenvironment, resulting in unique features in hematoxylin-eosin (HE) staining histopathological images6. DL methods have been proven efficient in recognizing complex feature patterns from HE images and contribute to personalized healthcare6.
Gastric cancer is one of the most common tumors in the world, with great histological diversity7,8 and molecular diversity9. The biological mechanisms and clinicopathologic characteristics of different gastric cancer subtypes are extremely different. Further gastric cancer classification is of great significance for diagnosis, treatment, monitoring, and prognosis. The histological gastric cancer subtypes have multiple classification methods, such as WHO classification, Lauren classification, and Japanese classification7. The classification of these histological subtypes is related to the adenocarcinoma differentiation grade, including well-differentiated adenocarcinoma (WDA) and poorly differentiated adenocarcinoma (PDA). The classification adenocarcinoma differentiation grade is based on the presence or absence of glandular structure formation10,11. The gastric cancer differentiation grade is usually closely related to the clinicopathologic characteristics and prognosis, and a well-differentiated tumor usually has a better prognosis and longer survival length12. In most of the reported studies, the adenocarcinoma differentiation grade is judged through manual identification by pathologists13. Many studies have developed DL systems to identify tumors14, but the problem of tumor differentiation grade recognition still needs further investigation.
Among the gastric cancer molecular subtypes, diagnosing microsatellite instability (MSI) status is of great importance for the treatment and prognosis of tumors9. Microsatellites are short tandemly repeated DNA sequences that are widely distributed throughout the human genome, accounting for ~3% of the genome15. Damage to the DNA mismatch repair (MMR) function can cause the insertion or deletion of microsatellites, leading to genetic mutations in MSI16. In the National Comprehensive Cancer Network (NCCN) guidelines, MSI status detection is an important part of the clinical diagnosis process17. Several MSI detection methods have made great progress, including immunohistochemistry (IHC), fluorescent multiplex polymerase chain reaction (PCR), and next-generation sequencing (NGS)18. All these MSI detection methods need to carry out additional diagnostic tests in addition to HE pathological staining, which requires more tissue samples, time, and costs6,14. To overcome these limitations, MSI detection methods based on HE staining images and DL technologies have attracted increasing attention19. Many studies have proven that DL models can recognize the MSI status from HE images14,20,21, but the relationships between MSI status and gastric tumor histological features are still unclear.
In this study, we established a DL system for differentiation grade recognition and MSI status recognition for gastric cancer. In addition, we used the convolutional neural network (CNN) visualization method to explore the key features extracted by the DL models in differentiation grade and MSI state recognition to improve DL system interpretability.
Materials and methods
Patient cohorts
We obtained patient samples with gastric cancer from the pathology archive at Beijing Cancer Hospital from 2015 to 2020. To increase the reliability of the dataset and DL models, we set strict criteria for screening the patient samples. The patients were archived based on the following criteria: (1) the pathological diagnosis was confirmed as gastric adenocarcinoma. In addition, according to the microscopic performance, the tumor was divided into two grades: well-differentiated (mainly composed of glandular tube-like structure) and poorly differentiated (unrecognizable or highly irregular adenoid structure; solid structure; scattered infiltration of poorly adherent tumor cells); (2) had undergone radical gastrectomy; and (3) the status of MSI was verified by IHC and PCR. If one or more of four major MMR proteins (MLH1, PMS2, MSH2, and MSH6) were identified to be lost, the sample was classified as mismatch repair deficient (dMMR). Due to the inconsistency between IHC and PCR results, cases with dMMR were further verified by PCR. The patients with dMMR/MSI-H tumors were classified as MSI type; otherwise, they were classified as MSS type.
Pathological HE staining images and MSI status information were collected for each patient. The digitized WSIs were acquired at Beijing Cancer Hospital using a Leica Aperio Versa pathologic scanner (scanned at ×40 magnification using a Leica ImageScope; maximum spatial resolution: 0.243 microns per pixel). Each WSI had ~109~1010 pixels. There were 467 cases in total. All these samples were divided into three cohorts: the training cohort with 348 annotated WSIs, the testing cohort with 88 annotated WSIs, and the integration testing cohort with 31 original WSIs without tumor contour annotation.
Annotation of the dataset
The annotation of the HE staining images comprises two sequential steps. We used Automated Slide Analysis Platform (ASAP) software to implement image annotation. First, we divided the tissues into tumor and nontumor regions. Furthermore, we labeled WDA and PDA regions in the tumor region. The whole annotation process was completed by two pathologists and finally reviewed by a senior pathologist.
Image tile extraction
Each WSI usually occupies up to gigabytes when uncompressed and cannot be directly processed by a computer22,23. We used the OpenSlide tool to segment the WSI into small image tiles for further analysis24,25. The original WSI was downsampled by a factor of two when extracting image tiles. The size of the tile was 512 × 512 pixels with a spatial resolution of 0.49 μm/pixel, and the edge length of the tile was 250 μm.
First, there was usually only a part of the region in the WSI that was valid with the presence of tissue samples, and the other invalid regions had RGB values of (0, 0, 0) in the mrxs format file. We abandoned the invalid region in the WSI by setting a threshold setting for the mean value of the RGB channels for each image tile. If the mean value for all channels is less than 200, the image tile is considered an invalid image tile. Otherwise, the image tile is valid and needs to be analyzed subsequently.
Next, we assigned labels to each image tile according to image annotations. In the tumor diagnosis task, the image tiles had three kinds of labels: other, PDA, and WDA. The tiles of the other class include muscle tissue, connective tissue, and normal glands. The rule to assign a label to an image tile: if an image tile belongs to label K, the area referring to label K should be greater than 75% of the total area of the image tile. In this study, we also used the term “tumor tile” to refer to the combination of PDA and WDA tiles. In the MSI diagnostic task, the tiles had two kinds of labels: MSS and MSI. Only tumor tiles were used in the MSI recognition task. The MSI status ground truth was patient-level labels. For MSI and MSS patients, all the corresponding tumor tiles were labeled MSI and MSS, respectively. Notably, color normalization was implemented over all the image tiles26.
DL model construction
For both tumor and MSI diagnostic tasks, all DL models were developed based on a pretrained ResNet-18 model based on ImageNet27. We used transfer learning techniques to fine-tune the DL models. Only the parameters in the last ten layers were updated, and the parameters in other layers were frozen. The image tiles were resized to 224 × 224 when input to the DL model. We used data augmentation methods to train the DL models, including random cropping and random flipping. In the DL model training process, the training image tiles were randomly shuffled, and 12.5% of the training tiles were used as the validation dataset. Other parameter settings in the training process were as follows: maximum epochs, 100; batch size, 256; learning rate, 10−6; L2 normalization, 10−4; optimizer, adaptive moment estimation (ADAM) algorithm. We checked the validation accuracy of the DL model on the validation dataset every 256 iterations. To avoid overfitting, the training process terminated if the accuracy was not improved for three consecutive validation times. The DL architectures and experiments were implemented on a computer with PyTorch and configured with an Nvidia GeForce RTX 2080 Ti GPU.
DL systems for tumor diagnosis
We divided tumor diagnosis into two sections: tumor recognition and tumor differentiation grade recognition. Tumor recognition denotes recognizing whether the input image tile belongs to the tumor class. Tumor differentiation grade recognition denoted not only recognizing the tumor but also distinguishing tumor differentiation grades. Corresponding to these two sections, we developed two models for tumor diagnosis. First, we established a two-class classification model for tumor recognition, which classified the input image tiles into tumor and other classes. Second, we established a three-class classification model for tumor differentiation grade recognition, which classified image tiles into PDA, WDA, and other classes. For the tumor differentiation grade recognition model, we combined the PDA and WDA recognition results as the tumor class and obtained the tumor recognition results.
DL systems for MSI diagnosis
MSI diagnosis included two adjacent modules: tile-level diagnosis based on DL models and patient-level diagnosis based on tile fusion. The tile-level diagnosis denotes using the DL models to calculate the MSS and MSI probability for each image tile and classify the tiles into MSI or MSS classes. In the patient-level MSI diagnostic process, we used two methods to fuse all tumor tile results from a patient and obtained the patient-level MSI status recognition results. The raw classifier directly compared the ratio of MSI tiles to the threshold of 50%, and the patients with more than 50% MSI tiles were classified as patient-level MSI cases. The optimal classifier explored the optimized ratio of MSI tiles using the training cohort to achieve better patient-level MSI diagnosis.
Evaluation of DL systems
The tumor diagnosis task includes tumor recognition using a two-class classification model and tumor differentiation degree recognition using a three-class classification model. We used general measures to evaluate all these models in the tumor diagnosis task. First, we used a confusion matrix to show the results of the models on the test dataset. In addition, we used precision, recall, and F1 score to evaluate the performance of different models in tumor diagnosis. Precision denotes how many of the predicted labels are actually in the ground truth. Recall denotes how many of the labels in the ground truth are correctly predicted. Both precision and recall have the best value of 1. However, precision and recall are sometimes contradictory. We used the F1 score, the weighted average of precision and recall, to comprehensively consider these two measurements.
where F1, P, and R denote the F1 value, precision, and recall, respectively.
Tumor MSI status recognition is a binary classification task. To ensure the effectiveness and reliability of the performance evaluation, we carry out both tile-level and patient-level MSI status recognition evaluations. We used the receiver operating characteristic (ROC) curve and area under the curve (AUC) to evaluate the tile-level performance of the models. The measurements of accuracy, the true negative rate (TNR), and the true positive rate (TPR) were used to evaluate the patient-level performance of the models. We adopted the bootstrap method to calculate the 95% confidence interval for patient-level accuracy. This study is implemented in adherence to Reporting of Diagnostic Accuracy Study (STARD) reporting guideline.
Gradient-weighted class activation map for DL models
One of the greatest problems for DL technology is model interpretability. DL models are usually regarded as a “black box”. We cannot understand how the DL models implement information processing and obtain the final results. Improving the intelligibility of DL models is very important, especially in the application of clinical diagnosis. The gradient-weighted class activation map (Grad-CAM) is a widely used CNN visualization method that can locate the critical area that the DL model focuses on in the decision-making process28,29. To validate the reliability and increase the interpretability of the DL system, we used the Grad-CAM method to show the key image features that the DL system extracted in the PDA/WDA and MSI/MSS classification process.
Results
Establishment of a DL system for tumor and MSI diagnosis
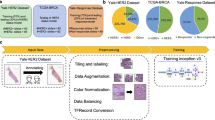
The DL system for tumor and MSI diagnosis comprises two sequential procedures: tumor diagnosis and MSI diagnosis (Fig. 1a). In the tumor diagnosis procedure, we used the sliding cropping method to acquire tiles from whole-slide images (WSIs). Then, we developed a two-class tumor recognition model to classify tiles into the other and tumor classes and a three-class tumor differentiation grade recognition model to classify tiles into the other, PDA, and WDA classes. In the following MSI diagnosis procedure, only the tumor tiles, including PDA and WDA, were considered. We developed MSI status recognition DL models to classify the tiles into MSS and MSI classes and then used a data fusion method to fuse all the tiles of a patient to obtain the patient-level MSI diagnosis result.
The architecture and functional relationships of the modules in the DL system are shown in Fig. 1b. The dataset module contains the original medical dataset (pathology WSIs and MSI information for each patient), region of interest (ROI) annotations for tumor, and image tile dataset extracted according to pathology WSIs and ROI annotation dataset. The deep learning module includes tumor diagnosis models and MSI diagnosis models. Human experts can directly annotate pathological images to obtain ROI annotation datasets and tile datasets or update these datasets based on the results of the tumor recognition model. In the application process, the DL system directly performs end-to-end diagnosis on WSIs and explores the key features extracted by the DL models in tumor differentiation grade and MSI status recognition through CNN visualization methods to improve system interpretability and reliability.
Performance of tumor diagnosis
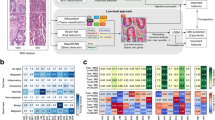
Tumor recognition is a fundamental task in pathological diagnosis, and further DL-based recognition of tumor differentiation grade will promote an accurate and personalized tumor diagnosis. We generated an image tile dataset according to the pathology WSI and image annotation dataset (Fig. 2a, b). These image tiles had three labels, including other, PDA, and WDA. Notably, the combination of the PDA and WDA tiles is called a tumor tile (Fig. 2b). We divided the patients into three classes according to the presence or absence of PDA and WDA tiles: patients with only PDA tiles (training: 117; testing: 42), patients with only WDA tiles (training: 107; testing: 46), and patients with both PDA and WDA tiles (training: 124; Fig. 2c, Table 1). To accurately evaluate the performance of DL systems in tumor differentiation grade recognition, the testing dataset contained patients with either PDA or WDA tiles. There were approximately one million image tiles in total, with approximately three hundred thousand tumor tiles. In both the training and testing datasets, the counts of the PDA and WDA tiles were well balanced (PDA:WDA; training, 1.15:1; testing, 1.03:1; Fig. 2d, Table 2).
a Annotations of PDA and WDA in WSI. Yellow, region of poorly differentiated adenocarcinoma (PDA). Red, region of well-differentiated adenocarcinoma (WDA). WSI, whole-slide image. Scale bar, 1 mm. b Samples of tiles for other, PDA, and WDA classes. The tumor class consists of PDA and WDA. Three tiles for each class are shown. Size of tiles, 512 × 512 pixels, 250 × 250 μm. c Pie graph of patient cohorts for tumor differentiation grade recognition. PDA and WDA denote the patients with only PDA and WDA tiles in the tumor class, respectively. PDA and WDA denote the patients with both PDA and WDA tiles. There are 436 patients in total. d Scatter plot for the number of tiles in different classes. Left panel, the plot of the tile count for PDA class versus the other class. Right panel, the plot of the tile count for the WDA class versus the other class. Each circle denotes a patient. Gray circles denote the training dataset. Red circles denote the testing dataset. e DL models to achieve tumor diagnosis. Left panel, the input image of an HE staining tile. Right panel, two DL models for tumor diagnosis, two-class (other/tumor) classification model, and three-class (other/PDA/WDA) classification model. The sample HE staining tile belongs to the tumor class and PDA class. f Heatmap of a confusion matrix for other/tumor classification. The number denotes the count of corresponding tiles. g Heatmap of a confusion matrix for other/PDA/WDA classification. h Gradient-weighted class activation map for PDA. Upper, original HE staining tiles for PDA class. Down, class activation maps for the HE staining tiles. Hot regions correspond to key features for DL models to recognize PDA. The red and blue colors indicate greater importance and less importance, respectively. i Gradient-weighted class activation map for WDA. Hot regions correspond to key features for DL models to recognize WDA.
To efficiently recognize the tumors and the tumor differentiation grade, we developed two models for tumor diagnosis, including a two-class classification model for tumor recognition and a three-class classification model for tumor differentiation grade recognition (Fig. 2e). In the tumor recognition task, the tiles in the testing dataset were greatly classified, and the F1 values for the other and tumor classes were 0.9699 and 0.9572, respectively (Fig. 2f, Table 3). In the tumor differentiation grade recognition task, the F1 values for the other, PDA, and WDA classes were 0.9741, 0.8615, and 0.8977, respectively. In addition, we used the combination of PDA and WDA to evaluate tumor recognition performance by a three-class classification model (F1 values for the other and tumor classes were 0.9741 and 0.9634; Fig. 2g, Table 3). In addition, key features that pathologists were concerned about were consistent with the features extracted by the DL model in identifying PDA and WDA tiles (Fig. 2h, i, Supplementary Fig. 1). The proposed DL system exhibited great tumor diagnosis performance and high interpretability.
Performance of MSI diagnosis
MSI was an important gastric cancer subtype. Identifying MSI status from pathological images using DL technologies will greatly increase the efficiency of clinical diagnosis and reduce costs. In the proposed DL system, we first used the DL model to perform MSS/MSI classification on tumor tiles. Then, the results of all tumor tiles from a patient were fused by the majority voting method to achieve patient-level MSI status recognition (Fig. 3a). There were 264 MSS patients (training: 212; testing: 52) and 172 MSI patients (training: 136; testing: 36) (Fig. 3b, Table 4). Among all the tumor image tiles (~3 × 105 tiles), there were 38.45% training MSI tiles, 30.63% training MSS tiles, 13.06% testing MSI tiles, and 17.86% testing MSS tiles (Fig. 3c, Table 5). We first implemented tile-level MSI recognition through the DL method and achieved an AUC of 0.7854 [95% CI 0.7825-0.7882] in the testing cohort (Fig. 3d). Furthermore, we fused the tile-level MSI results of all tumor tiles from a patient to achieve patient-level MSI status recognition (raw classifier with an accuracy of 77.27% [95% CI 68.57%–86.36%], true negative rate (TNR) of 69.23%, and true positive rate (TPR) of 88.89%; optimal classifier with an accuracy of 86.36% [95% CI 78.99–93.90%], TNR of 88.46%, and TPR of 83.33% in the testing cohort). (Fig. 3e). The optimal classifier exhibited the best performance in the testing cohort, and we used it as the tile fusion method in the following integration testing experiments.
a Overview of the MSI status recognition process by the DL system. Left to right, extracting tumor tiles from WSI, tile-level MSS/MSI classification, fusion of tile-level MSI results, and patient-level MSI status diagnosis. b Pie graph of patient cohorts for MSI status recognition. MSS and MSI denote the patients belonging to the MSS and MSI classes, respectively. There are 436 patients in total. c Pie graph of tumor tiles for MSI status recognition. MSS and MSI denote the tumor tiles belonging to the MSS and MSI classes, respectively. There are approximately three hundred thousand tumor tiles in total. d ROC curve plot for tile-level MSI recognition in testing cohorts. e Confusion matrix plot for patient-level MSI recognition in testing cohorts. The performance of the two methods are shown. Left panel, raw classifier comparing the ratio of MSI tiles to the threshold of 50%. Right panel, optimal classifier by comparing the ratio of MSI tiles to the threshold of 30.7%. f Gradient-weighted class activation map for MSS. Upper, original HE staining tiles for MSS tumor tiles. Down, class activation maps for the HE staining tiles. Hot regions correspond to key features for DL models to recognize MSS. The red and blue colors indicate greater importance and less importance, respectively. g Gradient-weighted class activation map for MSI. Hot regions correspond to key features for DL models to recognize MSI.
In addition, we used the Grad-CAM method to visualize the key features identified by the DL model in the MSI status recognition task (Fig. 3f, g, Supplementary Fig. 2). Although pathologists generally do not use HE staining images to diagnose MSI status, it is widely accepted that different MSI statuses usually correspond to specific pathological features. Our visualization results can promote establishing a clear relationship between MSI status and pathological characteristics and can also improve DL system interpretability and reliability.
Integration testing of the DL systems for tumor and MSI diagnosis
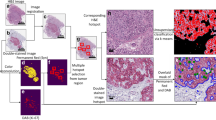
To further validate the effectiveness of the proposed DL system, we applied the integrated tumor differentiation grade and MSI status recognition models to the integration testing cohort of original WSIs without tumor contour annotation (17 MSS and 14 MSI cases) (Fig. 4). The system automatically segmented the WSI into small image tiles and implemented DL-based tumor differentiation grade recognition at the image tile level (Fig. 4a, b). The WDA and PDA tiles recognized by the system were marked on the original HE staining WSI to demonstrate the tumor diagnosis results. Furthermore, subsequent MSI status recognition was implemented for PDA and WDA tiles recognized by the system, and the MSI probabilities for all tumor tiles were exhibited using a heatmap (Fig. 4c). The integrated system used the optimal classifier for tile fusion and achieved automatic patient-level MSI diagnosis with an accuracy of 83.87%, TNR of 88.24%, and TPR of 78.57% in the integrated testing dataset. The proposed DL system integrated the tumor differentiation grade and MSI status recognition problems into the same workflow and was suitable for exploring the relationships between pathological features and molecular status.
a Original HE staining WSI. b Differentiation grade recognition based on HE stating WSI. WSI was segmented into image tiles to achieve differentiation grade recognition. Green squares denote WDA. Yellow squares denote PDA. c Heatmap of MSI probability for tumor tiles calculated by the MSI status recognition model. The tumor tiles consisted of WDA and PDA tiles in (b). d Patient-level MSI recognition in the integration testing dataset. Upper, the confusion matrix plot. Down, evaluation of patient-level MSI recognition performance. TNR true negative rate, TPR true positive rate.
Discussion
To overcome the limitations of the classic diagnostic procedure in gastric cancer, we established a DL system to achieve intelligent tumor differentiation grading and MSI status recognition from HE-stained WSIs. In addition, we used the CNN visualization method to demonstrate the key pathological features learned by the DL system to increase the interpretability of the system.
In the tumor evaluation process, it is necessary for pathologists to identify the degree of differentiation because well-differentiated tumors tend to have lower atypia, good adhesion, and an absence of metastasis; that is, they have a positive impact on guiding the choice of clinical treatment and predicting prognosis. Furthermore, previous research has shown that the 5-year survival rate and disease-free survival rate of intestinal-type gastric cancer are significantly different from those of diffuse-type and mixed-type gastric cancer30, so intestinal-type gastric cancer is related to a better prognosis. According to the corresponding relationship between histological grade and Lauren classification, that is, intestinal-type tumors correspond to high-moderate differentiation, well-differentiated tumors have a better prognosis. Therefore, we often regard the degree of differentiation as an independent risk factor affecting patient survival rate with gastric cancer in clinical work. Finally, we found that the detailed tumor differentiation grade recognition model exhibited better performance in recognizing tumor regions, which increases the accuracy of automatic tumor tile-based diagnosis, such as further MSI diagnosis.
Benefiting from advances in molecular biology research on molecular subtypes of tumors, such as MSI, great progress has been made. MSI was related to at least 14 kinds of cancers16. MSI is spatially popular in colorectal cancer, endometrial carcinoma, and gastric cancer, with an occurrence frequency of higher than 10%31. The diagnosis of MSI is of great significance for tumor treatment because most patients with MSI tumors have a good prognosis and better median overall survival32, and MSI tumors are especially suitable for immunotherapy18. Several MSI detection methods based on molecular assays have been well established. IHC detects the expression of MMR proteins, which can indirectly reflect the status of MSI. The loss of any MMR proteins composed of hMLH1, hPMS2, hMSH2, and hMSH6 indicates MMR deficiency (dMMR). Generally, dMMR is equivalent to MSI18. The PCR method compares the microsatellite loci detected in tumor tissue with normal DNA, which can directly reflect the MSI status and has high sensitivity and specificity33. With the development of genotyping technology, NGS has become an important tool for cancer genome analysis. NGS testing directly performs genome sequencing of known genes to test MSI status in tumor tissues34. In most tumor diagnostic processes, HE pathological staining is an indispensable test. However, these MSI detection methods require tests other than HE staining, which increases the time and cost of diagnosis6,14. Using deep learning methods to achieve MSI status recognition based on HE staining images can effectively solve these limitations. The DL and HE-based MSI detection methods provide an alternative to the molecular assay-based MSI detection method and will provide patients with fast and accurate cancer diagnosis services.
Most studies have reported excellent DL and HE-based MSI detection systems in colorectal cancer14,20,21,35. Kather et al. developed an MSI detection system from The Cancer Genome Atlas (TCGA) and Darmkrebs: Chancen der Verhütung durch Screening (DACHS) datasets; they adopted a two-step automatic tumor detection approach followed by subsequent MSI detection14. Yamashita et al. classified the HE WSI into seven different tissue types first and then predicted the MSI status only on the tumor-epithelial and mucinous tiles; the developed system showed higher performance than pathologists20. Echle et al. developed MSI detection models using a large international cohort with 6406 specimens from TCGA, DACHS, Quick and Simple and Reliable trial (QUASAR), and the Netherlands Cohort Study (NLCS) dataset; the prediction performance was robust in clinicopathologic and molecular subgroups such as different stages and locations21. Most of these reported studies developed DL models using supervised learning techniques. Bilal et al. developed a weakly supervised DL framework to predict key mutations in colorectal cancer from HE images; they also predicted the status of molecular pathways by characterizing histological features of different cellular compositions35.
Multiple reported studies have also achieved MSI detection from HE images in gastric cancer36,37,38. The most recently reported study developed DL models to detect MSI and Epstein–Barr virus (EBV) status using ten patient cohorts with gastric cancer from seven countries; different subgroups (including various UICC stages, Lauren histological subtypes and differentiation grades) showed subgroup-dependent performance in MSI and EBV detection36. In this study, we achieved automatic tumor differentiation grade and MSI status recognition in gastric cancer. Furthermore, the three-class classification model (other/PDA/WDA) showed better performance than the two-class classification model (other/tumor) in tumor recognition tasks, providing a better foundation for predicting MSI status. We also exhibited the key pathological features related to the tumor differentiation grade and MSI status learned by DL models using the CNN visualization method. Compared to Muti et al.’s patch-level visualization method36, our work exhibited more detailed pathological features and increased the interpretability of the models. Apart from model differences, we also collected a large WSI dataset with 467 cases in our study. We used 74.52% (348/467) of the cohorts to train the DL models, 18.84% (88/467) of the cohorts to test the effectiveness of the tumor differentiation model and MSI detection model, and the remaining 6.64% (31/467) of the cohorts to test the MSI detection performance of the integrated system. Compared with most of reported studies14,20, the training and testing cohorts in this study are large enough to ensure the reliability of DL models. Muti et al. included 2823 patients in ten cohorts from seven countries. Large-scale and multicenter validation make their work more general and have great potential for incorporation into clinical workflows36. However, Muti et al.’s study involved no Chinese patients, and this study implemented complementary studies on Chinese patients.
Clarifying the relationships between MSI status and histological features for gastric cancer will not only provide insight into the mechanism of its development and treatment but also increase the reliability of DL-driven MSI status recognition methods. In contrast to colorectal cancer, in which MSI is related to mucinous differentiation, poor differentiation, Crohn’s-like reaction and tumor budding20, gastric cancer with MSI is associated with well-differentiated morphological features39. Consequently, it is meaningful for us to use deep learning to study the MSI status of gastric cancer directly from HE slides.
To increase the interpretability of the DL system, we used the CNN visualization method, Grad-CAM, to exhibit the key features learned by DL models. The main pathological features of adenocarcinoma include larger cells, larger nuclei, and a higher nucleus/cytoplasm ratio. The key pathological features of the formation of glandular structures to distinguish PDA and WDA were successfully extracted by DL models. These experimental results further confirmed the correctness and reliability of DL models in tumor diagnosis based on HE images. We further extracted the key pathological features in the MSS/MSI classification process. The CNN visualization method will be a great tool for establishing the mapping between morphological characteristics of gastric cancer and MSI subtypes. In addition, the number of cases included in our study has been expanded compared with previous studies, making our study more credible in identifying MSI morphological characteristics and further applying it to clinical screening.
The main limitation of this study lies in the lack of tile-level ground truth in the MSI diagnosis task. The ground truth for MSI status was patient-level. In the MSI recognition model training, we assumed that all image tiles from a patient had the same MSI status. However, tumor tissues usually have great heterogeneity, and even MSI patients will have MSS tumor tiles. In our further study, we plan to use weakly supervised learning techniques, such as multi-instance learning, to develop the MSI status recognition model to eliminate the impact of the lacking tile-level ground truth.
Another limitation is that the pathological subtypes considered in this study are not rich enough. The histological differentiation grade was correlated with the Lauren classification. It is generally accepted that well- and poorly-differentiated tumors correspond to intestinal and diffuse types. Identification of tumor differentiation status plays an important role in clinical treatment selection and prognosis prediction. In addition, gastric cancer has several pathological subtypes according to the WHO classification, such as tubular adenocarcinoma, papillary adenocarcinoma, and signet-ring-cell carcinoma. There are usually some overlaps between signet-ring-cell carcinomas and poorly differentiated tumors, and the signet-ring-cell carcinoma identification is also essential for precise diagnosis and treatment40. Tumor differentiation grade recognition and tumor subtype recognition are both critical to achieving personalized medicine. In this study, we focused on the recognition of tumor differentiation grade and MSI status. In our future research, we will do our best to expand the patient samples, incorporate more pathological subtypes, such as signet-ring-cell carcinomas, and improve the clinical utility of the system.
In conclusion, the proposed DL system is end-to-end and has integrated multiple clinical diagnosis procedures, including tumor differentiation grade and MSI status recognition. The system has high accuracy, reliability, and interpretability and is strongly generalizable to other clinical diagnosis tasks. Thus, the system has great potential to be integrated into clinician workflows and provides a step forward in the implementation of artificial intelligence healthcare.
Data availability
The dataset was collected and held by the Beijing Cancer Hospital. Annotated image tile datasets are available for noncommercial use at https://zenodo.org/record/5155995#.YQoeccgl3RR. Other datasets analyzed during the current study are available from the corresponding author upon reasonable request.
Code availability
Source codes are available at https://github.com/Benson0704/MSI_MSS_Prediction.
References
Esteva, A. et al. A guide to deep learning in healthcare. Nat. Med. 25, 24–29 (2019).
Norgeot, B., Glicksberg, B. S. & Butte, A. J. A call for deep-learning healthcare. Nat. Med. 25, 14–15 (2019).
Su, F. et al. Development and validation of a deep learning system for ascites cytopathology interpretation. Gastric Cancer 23, 1041–1050 (2020).
Kather, J. N. et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 16, e1002730 (2019).
Hu, Y. et al. Deep learning system for lymph nodes quantification and metastatic cancer identification from whole-slide pathology images. Gastric Cancer 24, 868–877 (2021).
Kather, J. N. & Calderaro, J. Development of AI-based pathology biomarkers in gastrointestinal and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 17, 591–592 (2020).
Arai, T. et al. Frequent microsatellite instability in papillary and solid-type, poorly differentiated adenocarcinomas of the stomach. Gastric Cancer 16, 505–512 (2013).
Sugimura, H. Editorial: an obsession with subtyping gastric cancer. Gastric Cancer 16, 451–453 (2013). vol.
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014).
Kanesaka, T. et al. Clinical predictors of histologic type of gastric cancer. Gastrointest. Endosc. 87, 1014–1022 (2017).
Kuwata, T. et al. Establishment of novel gastric cancer patient-derived xenografts and cell lines: pathological comparison between primary tumor, patient-derived, and cell-line derived xenografts. Cells 8, 585 (2019).
Feng, F. et al. Prognostic value of differentiation status in gastric cancer. BMC Cancer 18, 865 (2018).
Liu, S. et al. Apparent diffusion coefficient value of gastric cancer by diffusion-weighted imaging: Correlations with the histological differentiation and Lauren classification. Eur. J. Radiol. 83, 2122–2128 (2014).
Kather, J. N. et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 25, 1054–1056 (2019).
Cortes-Ciriano, I., Lee, S., Park, W.-Y., Kim, T.-M. & Park, P. J. A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun. 8, 15180 (2017).
Hause, R. J., Pritchard, C. C., Shendure, J. & Salipante, S. J. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 22, 1342–1350 (2016).
Messersmith, W. A. NCCN guidelines updates: management of metastatic colorectal cancer. J. Natl. Compr. Cancer Netw. 17, 599–601 (2019). vol.
Li, K., Luo, H., Huang, L., Luo, H. & Zhu, X. Microsatellite instability: a review of what the oncologist should know. Cancer Cell Int. 20, 16 (2020).
Hildebrand, L. A., Pierce, C. J., Dennis, M., Paracha, M. & Maoz, A. Artificial intelligence for histology-based detection of microsatellite instability and prediction of response to immunotherapy in colorectal cancer. Cancers. 13, 391 (2021).
Yamashita, R. et al. Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet. Oncol. 22, 132–141 (2021).
Echle, A. et al. Clinical-grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology 159, 1406–1416 (2020). e11.
Pantanowitz, L. et al. Twenty years of digital pathology: an overview of the road travelled, what is on the horizon, and the emergence of vendor-neutral archives. J. Pathol. Inform. 9, 40 (2018).
Herrmann, M. D. et al. Implementing the DICOM standard for digital pathology. J. Pathol. Inform. 9, 37 (2018).
Goode, A., Gilbert, B., Harkes, J., Jukic, D. & Satyanarayanan, M. OpenSlide: a vendor-neutral software foundation for digital pathology. J. Pathol. Inform. 4, 27 (2013).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Macenko, M., et al. A method for normalizing histology slides for quantitative analysis. in 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro 1107–1110 (2009).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. in Proceedings of the IEEE conference on computer vision and pattern recognition 770–778 (2016).
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A. & Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE conference on computer vision and pattern recognition 2921–2929 (2016).
Selvaraju, R. R. et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 128, 336–359 (2020).
Chen, Y.-C. et al. Clinicopathological variation of lauren classification in gastric cancer. Pathol. Oncol. Res. 22, 197–202 (2016).
Baretti, M. & Le, D. T. DNA mismatch repair in cancer. Pharmacol. Ther. 189, 45–62 (2018).
Fan, J. P., Qian, J. & Zhao, Y. J. The loss of PTEN expression and microsatellite stability (MSS) were predictors of unfavorable prognosis in gastric cancer (GC). Neoplasma 67, 1359–1366 (2020).
Suraweera, N. et al. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology 123, 1804–1811 (2002).
Hempelmann, J. A. et al. Microsatellite instability in prostate cancer by PCR or next-generation sequencing. J. Immunother. cancer 6, 29 (2018).
Bilal, M., et al. Development and validation of a weakly supervised deep learning framework to predict the status of molecular pathways and key mutations in colorectal cancer from routine histology images: a retrospective study. Lancet. Digit. Heal. (2021) https://doi.org/10.1016/S2589-7500(21)00180-1.
Muti, H. S. et al. Development and validation of deep learning classifiers to detect Epstein-Barr virus and microsatellite instability status in gastric cancer: a retrospective multicentre cohort study. Lancet. Digit. Heal 3, e654–e664 (2021).
Chen, Y. et al. The immune subtypes and landscape of gastric cancer and to predict based on the whole-slide images using deep learning. Front. Immunol. 12, 685992 (2021).
Hinata, M. & Ushiku, T. Detecting immunotherapy-sensitive subtype in gastric cancer using histologic image-based deep learning. Sci. Rep. 11, 22636 (2021).
Ratti, M., Lampis, A., Hahne, J. C., Passalacqua, R. & Valeri, N. Microsatellite instability in gastric cancer: molecular bases, clinical perspectives, and new treatment approaches. Cell. Mol. Life Sci. 75, 4151–4162 (2018).
Zhang, S. et al. REUR: a unified deep framework for signet ring cell detection in low-resolution pathological images. Comput. Biol. Med. 136, 104711 (2021).
Funding
This work was supported by the National Key Research and Development Program of China (2018YFC0910700), the Major Program of National Natural Science Foundation of China (91959205).
Author information
Authors and Affiliations
Contributions
Conceptualization: Y.S., J.-F.J.; Methodology: F.S., J.L., X.Z., and B.W.; Formal analysis and investigation: F.S., J.L., X.Z., B.W., Y.H., and Y.S.; Writing, review, and/or revision of the manuscript: F.S., J.L., X.Z., B.W., Y.H., Y.S., and J.-F.J.; Funding acquisition: JL, YS; Resources: Y.S., J.-F.J.; Supervision: Y.S., J.L., J.-F.J.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics Approval/Consent to Participate
The study was performed in accordance with relevant guidelines and regulations and approved by the institutional review board at Beijing Cancer Hospital. All patients provided written informed consent to participate in the institutional review board-approved protocol.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Su, F., Li, J., Zhao, X. et al. Interpretable tumor differentiation grade and microsatellite instability recognition in gastric cancer using deep learning. Lab Invest 102, 641–649 (2022). https://doi.org/10.1038/s41374-022-00742-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-022-00742-6