Abstract
DNA methyltransferase 1 (DNMT1) shows close link with heart disease. This study aimed to define the role DNMT1 plays in heart failure and determine the underlying mechanism. Expression of microRNA (miR)-152-3p, DNMT1, E26 transformation specific-1 (ETS1) and ras homolog gene family member H (RhoH) was determined by RT-qPCR and/or western blot analysis. The interaction between miR-152-3p and ETS1 was predicted and verified. Methylation of the miR-152-3p promoter region was assessed using methylation-specific PCR. H9c2 cells were chosen for in vitro assays to examine the regulatory role of DNMT1 in autophagy and mitophagy with respect to miR-152-3p/ETS1/RhoH. Doxorubicin (DOX)-induced rat models of heart failure were employed for in vivo validation. DNMT1 expression was upregulated in the heart tissues of DOX-induced rats, where it showed an inverse correlation with miR-152-3p expression. Moreover, DNMT1 was shown to enhance methylation of the miR-152-3p promoter region and suppress its expression, leading to inhibition of mitophagy in H9c2 cells. In addition, DNMT1 enhanced expression of ETS1, which further elevated RhoH expression. Moreover, ETS1-elevated RhoH reduced cell viability and promoted autophagy and mitophagy in H9c2 cells upon treatment with DOX. Next, in vivo results demonstrated that depletion of DNMT1 protected rats from heart failure in a miR-152-3p/ETS1/RhoH-dependent manner. Overall, these findings indicate that DNMT1 may inhibit expression of miR-152-3p by promoting the methylation of miR-152-3p and enhancing the expression of ETS1, thereby inducing RHOH transcriptional activation and inhibiting mitochondrial autophagy, ultimately promoting the development of heart failure.
Similar content being viewed by others
Introduction
Heart failure has become a rapidly increasing global health issue that affects more than 37.7 million individuals worldwide1. With the increase in the population over the age of 65 years old, the prevalence of heart failure is increasing during this decade2. Therefore, a better understanding of this complicated pathophysiological syndrome will improve therapeutic outcomes and reduce the socioeconomic burden.
DNA methyltransferase 1 (DNMT1), whose primary function is to restore methylation to newly synthesized daughter DNA strands3, plays critical roles in the development of smooth muscle cells4. DNA methylation is an epigenetic modification that is essential for normal mammalian development, and abnormal DNA methylation is associated with a variety of diseases, including heart failure5,6. Expression of DNMT1 is rapidly upregulated in heart tissues of pressure overload- and adriamycin-induced heart failure mice, consistent with the increased expression of DNMT1 observed in familial hypertrophic cardiomyopathy patients7. Meanwhile, expression of DNMT1 has also been found to be increased in hyperhomocysteinemia-induced heart failure8. Interestingly, a recent study has demonstrated that depletion of DNMT1 protects the myocardium from heart failure, yet the underlying mechanism has not been elucidated7. An increasing body of evidence has shown that DNMT1 suppresses the expression of microRNA (miR)-152-3p in a methyltransferase activity-dependent manner9,10. As previously reported, upregulation of miR-152-3p exerts a protective role in cardiomyocytes11. On the other hand, E26 transformation specific-1 (ETS1), a transcription factor, exerts essential functions in embryogenesis, angiogenesis and inflammation and is involved in angiotensin-II-induced cardiac fibrosis, which contributes to the pathogenesis of heart failure12. The STRING database predicted the presence of an interaction between ETS1 and ras homolog gene family member H (RhoH). RhoH is an atypical guanosine triphosphatase (GTPase) that lacks GTPase activity and remains in the active conformation, which has been implicated in human diseases13. Of note, inhibition of the RhoA/Rho kinase signaling pathway ameliorates adriamycin-induced cardiac damage and DNA damage, restores heart function, and suppresses apoptosis and cellular senescence14. Moreover, pharmacological inhibition of Rho-associated kinases prevents cardiac fibrosis, which is a hallmark of heart failure, by suppressing cardiac myofibroblast function15. Rho kinase inhibition has been reported to improve mitochondrial function during liver injury16. Mitophagy represents a mitochondrial quality control mechanism through which damaged or excess mitochondria are eliminated to maintain cardiac function in response to varied stress and cardiac disease conditions17. Dysregulated mitophagy has been linked to the progression of heart failure and aging18. These findings suggest that ETS1/RhoH may suppress mitophagy, exaggerate heart failure and favor worse clinical outcomes. Therefore, targeting the ETS1/RhoH axis could be a potential therapeutic approach. However, whether ETS1 and RhoH are involved in the progression of DNMT1/miR-152-3p-mediated heart failure initiation/progression is unclear. In this study, we aimed to elucidate the role of DNMT1 in the pathogenesis of heart failure and define the underlying molecular mechanism. We investigated whether DNMT1-regulated miR-152-3p and ETS1/RhoH were associated with the pathological process of heart failure.
Materials and methods
Ethics statement
The current study was approved by the Animal Ethics Committee of the Third Hospital of Hebei Medical University. We attempted to reduce the suffering of the animal used during the experiments.
Microarray-based gene expression profiling
The heart failure-related gene expression dataset GSE84796 (Homo sapiens) was retrieved from the Gene Expression Omnibus (GEO) database, which contained seven normal samples and ten heart failure samples. Differential analysis was conducted using the R language “limma” package with |logFC | >0.8 and false discovery rate (FDR) < 0.05 as the threshold (the FDR method was used to correct the difference in p value) to obtain differentially expressed genes related to heart failure. The STRING database was applied to predict potential genes interacting with RhoH. Correlation analysis between RhoH and the predicted interacting gene ETS1 was performed using the ChIPBase database. The methylation site of the miR-152-3p promoter was predicted using the MethPrimer database. Meanwhile, the GeneMANIA database was used for correlation analysis between targets and disease function.
Establishing rat models of heart failure
Rat heart failure models were established as previously reported19,20. Briefly, healthy Sprague–Dawley rats (n = 50; 6–8 weeks old, weighing 220–260 g, Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were used. Doxorubicin (DOX) was prepared in a 2.0 mg/mL solution with normal saline. Ten rats were intraperitoneally injected with DOX solution at a dose of 2 mL/kg body weight once a week for 6 consecutive weeks, for a total of 24 mg/kg. In parallel, ten rats in the control group received the same amount of normal saline. The amount of DOX was determined based on the rats’ body weight before each injection.
A total of 50 Sprague–Dawley rats were randomly divided into control (n = 10) and DOX (DOX-induced heart failure models) groups. Rats in the DOX groups were further randomly divided into 3 groups: sh-NC + LV-NC + DOX (n = 10), sh-DNMT1 + LV-NC + DOX (n = 10) and sh-DNMT1 + LV-RHOH + DOX (n = 10). pLV-EGFP-N (gene overexpression vector) and pSIH1-H1-copGFP (gene silencing vector) lentiviral vectors were used for overexpression and silencing of genes. LV-NC, LV-RHOH, sh-NC, and sh-DNMT1 recombinant lentiviral particles and virus packaging kits were purchased from Shanghai Genechem Co., Ltd. (Shanghai, China). After 48 h of co-transfection of HEK293T cells with lentiviral transfection reagents, the viral titer was measured using a p24 ELISA kit (Cell Biolabs, Inc., San Diego, USA), and the virus was titrated to 109 TU/mL. The constructed and packaged lentivirus was then injected into rats in different groups via the tail vein. Each rat was injected with 100 μL virus solution (108 TU). After 72 h, relevant detection and observation were performed.
Echocardiography
After the rats were anesthetized, they were fixed in a supine position. The probe was placed at the left edge of the sternum. The maximum diameter of the long axis of the left ventricle was selected, and M-mode ultrasound was used to mark the end systolic and end diastolic endocardium of the ventricular septum and left ventricular wall. The left ventricular ejection fraction (LVEF), left ventricular shortening fraction, left ventricular systolic diameter (LVSD), and left ventricular diastolic diameter (LVDD) were automatically calculated using random software, with five cardiac cycles selected and averaged.
Pathological observation
After euthanasia, heart tissues were collected, washed with PBS and weighed. The ventricle tissues were fixed in 4% paraformaldehyde, embedded in paraffin and cut into 5 μm-thick sections. The sections were then submitted to hematoxylin eosin (H&E) staining. Next, two pathologists blinded to this study performed the pathological evaluation.
Mitochondrial isolation
Isolation of mitochondria was performed based on a previously published method21. Isolated mitochondria were incubated on wet ice for 15 min followed by measurement of protein concentration using the Bradford method with bovine serum albumin as a standard. All steps were performed on wet ice.
In-gel ATPase activity assay
Blue native PAGE (BN-PAGE) was employed to resolve natural and complete mitochondrial protein complexes. Upon completion of BN-PAGE, the gels were incubated in assay buffer (35 mM Tris/Cl, 270 mM glycine, 14 mM MgSO4, 0.2% (w/v) Pb(NO3)2, 8 mM ATP, pH 8.3) for the indicated periods, fixed in 50% methanol (incubation time) and rinsed twice in deionized distilled water (10 min each) followed by gel scanning. The precipitated lead from each sample was quantified utilizing densitometry.
Mitochondria membrane potential assay
Determination of mitochondrial membrane potential was performed utilizing a JC-1 Kit (GeneCopoeia, Rockville, MD, USA). Generally, mitochondria were isolated from heart tissues and treated with 5 μM JC-1 reagent for 10 min. The relative fluorescence was measured in a Turner microplate reader (Infinite 200, Tecan) under 485/535 nm excitation and 535/595 nm emission.
Cell culture and transfection
The H9c2 rat myoblast cell line (Procell, China) and HEK293T cells (CBP60439, COBIOER BIOSCIENCES CO., LTD., Nanjing, Jiangsu, China) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% (w/v) fetal bovine serum, 100 U/L penicillin and 100 mg/mL streptomycin. Cells were maintained in a 37 °C and 5% CO2 environment. Cell passage was performed every 2–3 days. Cell transfection was performed using Lipofectamine 2000 transfection reagent (Invitrogen, USA) with DNMT1 overexpression (oe-DNMT1), DNMT1 knockdown (sh-DNMT1), miR-152-3p mimic, miR-152-3p inhibitor, oe-ETS1, or sh-RhoH as well as the related controls (oe-NC, sh-NC, mimic-NC, inhibitor NC) or treated with DNA methylase inhibitor 5-azacytidine (5-Aza). All related plasmids were purchased from GeneChem (Shanghai, China).
For construction of Parkin-KO and ATG7-KO cell lines, Parkin and ATG7 gRNA were designed using tools.genome-engineering.org to generate a LentiCRISPR lentivirus CRISPR/Cas9 vector. In short, lentivirus (cloned sgRNA) was cotransfected with packaging plasmids pVSVg (AddGene 8454) and psPAX2 (AddGene 12260) into HEK293T cells. The packaged lentivirus was used to transduce the target cells, and positive clones were screened using puromycin. Parkin-KO (H9c2 cells with parkin knockout) and ATG7-KO cell lines (H9c2 cells with ATG7 knockout) were verified by western blot analysis and used for subsequent experiments.
Assessment of cell viability
Cell viability was assessed using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and lactate dehydrogenase (LDH) release assay as previously reported22. The spectrophotometric absorbance of the samples at 570 nm was measured using a microplate reader (Thermo Fisher 1510). LDH release was measured using a commercially available kit (Jiancheng Bioengineering Institute, Nanjing, China). The absorbance at 340 nm was measured using a microplate reader, and relative LDH release is presented as the percentage of LDH in the medium vs. the total LDH activity in cells.
Autophagy analysis by monodansylcadaverine (MDC) staining
Cells were fixed in 4% paraformaldehyde for 10 min at room temperature and stained with MDC (50 μmol/L) in PBS for 30 min at 37 °C. After completion of staining, the cells were monitored under an inverted fluorescence microscope. The cells were then suspended in PBS and analyzed in a flow cytometer. The relative content of autophagic vacuoles in the cells was reflected by the intensity of fluorescence. Data were analyzed using Win MDI2.9 software.
Dual-luciferase reporter assay
The wild-type (WT) and mutated 3′untranslated region (3′UTR) of ETS1 containing the predicted miR-152-3p targeting sequence were separately cloned into the psiCheck2 plasmid, and the generated plasmids were called EST-WT and EST-mutant type (MUT), respectively. Cells were cotransfected with EST-WT or EST-MUT and miR-152-3p mimic or miR-152-3p inhibitor. Luciferase activity was measured utilizing the Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI).
Methylation-specific polymerase chain reaction (MSP)
The methylation status of genomic DNA was evaluated using MSP as previously reported23. In brief, genomic DNA was extracted using a DNAMaxi Kit (Qiagen), and DNA was modified with sodium bisulfite using a Methyl Code Bisulfite Conversion Kit (Invitrogen). The primers used for detection have been reported in a previous study23.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using an RNeasy Mini Kit (Qiagen, Germany). Complementary DNA (cDNA) was synthesized using a PrimeScript RT reagent Kit (RR047A, Takara, Tokyo, Japan) or miRNA First Strand cDNA Synthesis (tailing Reaction) kit (B532451-0200, Sangon Biotech, Shanghai, China) to evaluate mRNA or miRNA expression levels, respectively. Relative expression of DNMT1, ETS1, RhoH and miR-152-3p was determined using a SYBR Premix Ex TaqTM II (Perfect Real Time) Kit (DDR081, Takara) on an ABI 7500 instrument (Applied Biosystems, Foster City, CA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 were used as internal controls for mRNA and miRNA quantification, respectively. Primer sequences are listed in Supplemental Table 1.
Western blot analysis
Total protein extraction from heart tissues or cells was performed using radioimmunoprecipitation assay lysis buffer (R0010, Solarbio, Beijing, China) supplemented with phenylmethylsulfonyl fluoride, and the concentration was determined using a bicinchoninic acid assay kit (23225, Pierce, Paris, France). Protein samples (50 μg/sample) were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (P0012A, Beyotime, Shanghai, China) and transferred onto polyvinylidene fluoride membranes. The membranes were blocked in 5% skimmed milk and then incubated with the following primary antibodies (all from Abcam, Cambridge, UK) overnight at 4 °C: DNMT1 (ab188453, 1:1,000), ETS1 (ab220361, 1:1,000), RhoH (ab118507, 1:1,000), Beclin-1 (ab207612, 1:1,000), LC3B (ab192890, 1:1,000), P62 (ab240635, 1:1,000), ATPIF1 (ab110277, 1:1,000), ATG7 (ab133528, 1:1,000), parkin (ab77924, 1:1,000), and β-actin (ab179467, 1:1,000). The next day, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) (Zhongshan Biotech, Guangzhou, China; 1:5,000). The immunocomplexes on the membrane were visualized using enhanced chemiluminescence reagent. Relative protein levels were quantified using Quantity One v4.6.2 software with β-actin as a loading control.
Statistical analysis
All data were analyzed using SPSS 21.0 software (IBM Corp., Armonk, NY, USA). Measurement data are presented as the mean ± standard deviation. Data between two groups were compared using unpaired t-test, and differences among multiple groups were analyzed by one-way analysis of variance followed by Tukey’s multiple comparisons test. Pearson’s correlation analysis was used to analyze the correlation between miR-152-3p and DNMT1 or ETS1 or between RhoH and ETS1. A value of p < 0.05 was considered statistically significant. All experiments were repeated three times.
Results
DNMT1 is highly expressed while miR-152-3p is poorly expressed in the rat model of heart failure
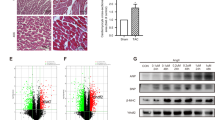
DNMT1 is highly expressed in homocysteine-induced heart failure8. A previous study reported that DNMT1 could bind to the miR-152-3p promoter to induce DNA methylation and inhibit the expression of miR-152-3p23. Moreover, upregulation of miR-152-3p alleviates heart failure24, but whether this is related to the regulation of DNMT1 remains unclear. To elucidate whether DNMT1 mediates miR-152-3p in the regulation of heart failure, we first established a rat heart failure model induced by DOX. The echocardiography results showed that DOX-treated rats displayed decreased LVEF and LVFS and elevated LVDD and LVSD (Supplemental Table 2). Moreover, H&E staining results of rat heart tissues revealed that in the control group, the architectural arrangement of the myoblasts remained parallel, and the nuclei were clear. Similarly, cellular edema and inflammatory cell infiltration were not observed. However, in the DOX group, tissue damage was observed, as reflected by cellular edema, hydropic degeneration, increased cellular distance between myocytes and infiltration of immune cells, including neutrophils, monocytes, and lymphocytes (Fig. 1A). These observations indicated successful establishment of the rat heart failure model.
A Representative H&E images displaying the histological characteristics of heart tissues from control rats and DOX-treated rats. Scale bar = 50 μm. Arrowhead indicates neutrophils. B Relative expression of DNMT1 in heart tissues from control rats and DOX-treated rats determined by RT-qPCR. C DNMT1 protein expression in heart tissues from control rats and DOX-treated rats determined by western blot analysis. D Expression of miR-152-3p in heart tissues from control rats and DOX-treated rats determined by RT-qPCR. E Pearson’s correlation analysis evaluating the relationship between DNMT1 expression and miR-152-3p expression in heart tissues from DOX-treated rats. *p < 0.05, compared to control rats. n = 10 rats in each group.
In addition, we observed higher DNMT1 but reduced miR-152-3p levels in heart tissues of DOX-treated rats (Fig. 1B–D, Supplemental Fig. 1A). Furthermore, Pearson’s correlation analysis indicated that the expression of DNMT1 was inversely correlated with that of miR-152-3p in the heart tissues of DOX-treated rats (Fig. 1E).
DNMT1 inhibits the expression of miR-152-3p by increasing DNA methylation of the miR-152-3p promoter region
We next determined how DNMT1 affects miR-152-3p during heart failure. The MethPrimer database predicted the presence of CpG islands in the promoter region of miR-152-3p (Fig. 2A). Therefore, we speculated that DNMT1 affected the occurrence of heart failure by regulating the methylation of miR-152-3p. MSP results showed that the methylation rate of CpG islands in the miR-152-3p promoter region was significantly higher in heart tissues of DOX-treated rats than in control rats (Fig. 2B, C). In addition, transfection with oe-DNMT1 increased the expression of DNMT1 in H9c2 cells, while there was no significant alteration in the expression of DNMT1 in H9c2 cells in response to treatment with oe-DNMT1 + 5-Aza (Fig. 2D, E, Supplemental Fig. 1B). RT-qPCR results showed that expression of miR-152-3p was decreased in H9c2 cells overexpressing DNMT1; however, when we treated DNMT1-overexpressing H9c2 cells with 5-Aza, a DNA methyltransferase inhibitor25, the inhibitory effect of DNMT1 overexpression on miR-152-3p expression was diminished (Fig. 2F). Furthermore, MSP assay results demonstrated that DNMT1 overexpression increased the methylation levels of miR-152-3p, while further 5-Aza treatment reversed this increase (Fig. 2G). These findings suggested that DNMT1 might inhibit the expression of miR-152-3p by enhancing DNA methylation at its promoter region.
A Methylation site of miR-152-3p promoter predicted by the MethPrimer database. B, C MSP assay detecting DNA methylation at the miR-152-3p promoter region using heart tissues from normal and DOX-treated rats. U indicates unmethylated status, and M indicates methylated status. D DNMT1 expression in H9c2 cells treated with oe-DNMT1 alone or in combination with 5-Aza was confirmed by RT-qPCR. E DNMT1 protein expression in H9c2 cells treated with oe-DNMT1 or combined with 5-Aza determined by western blot analysis. F Expression of miR-152-3p in H9c2 cells treated with oe-DNMT1 or combined with 5-Aza determined by RT-qPCR. G The methylation status of the miR-152-3p promoter region in H9c2 cells treated with oe-DNMT1 or combined with 5-Aza was evaluated by MSP assay. *p < 0.05, compared to control rats or H9c2 cells transfected with oe-NC. #p < 0.05, compared to H9c2 cells transfected with oe-DNMT1. n = 10 rats in each group. The cell experiments were independently conducted three times.
DNMT1 suppresses mitophagy of H9c2 cells by inhibiting miR-152-3p expression
The role of DNMT1 and miR-152-3p in mitophagy in H9c2 cells was our next focus. RT-qPCR data showed that oe-DNMT1 elevated the expression of DNMT1 while decreasing that of miR-152-3p in H9c2 cells. However, the miR-152-3p mimic did not alter the expression of DNMT1 but upregulated that of miR-152-3p. In addition, combined treatment with oe-DNMT1 and miR-152-3p mimic led to much higher miR-152-3p expression relative to oe-DNMT1 alone (Fig. 3A).
H9c2 myoblasts were treated with oe-DNMT1, miR-152-3p mimic or both. A DNMT1 and miR-152-3p in myoblasts determined by RT-qPCR. B, C Results of the MTT assay and LDH release assay evaluating viability and cell injury in myoblasts. D MDC-positive myoblasts, scale bar = 25 μm. E Protein expression of DNMT1, Beclin-1, P62, LC3-II, and LC3-I in cells determined by western blot analysis. *p < 0.05, compared to control cells or cells treated with oe-NC + mimic-NC; #p < 0.05, compared to cells treated with oe-NC + mimic NC + DOX; &p < 0.05, compared to cells treated with oe-DNMT1 + mimic-NC + DOX. The cell experiments were independently conducted three times.
Moreover, we found that DOX inhibited H9c2 cell viability and enhanced the LDH release rate in the cell culture solution, indicating increased H9c2 cell injury. DNMT1 overexpression aggravated H9c2 cell injury, which was decreased following miR-152-3p overexpression. Conversely, combined treatment with oe-DNMT1 and miR-152-3p mimic reversed the promoting effect of oe-DNMT1 on H9c2 cell injury (Fig. 3B, C). MDC staining results revealed that the fluorescence intensity and MDC-positive cells were both decreased in cells treated with DOX, suggesting the accumulation of autophagic vacuoles in DOX-treated cells and inhibition of autophagy. In contrast, autophagy was decreased in cells overexpressing DNMT1 but was enhanced upon miR-152-3p overexpression. In addition, combined treatment with oe-DNMT1 and miR-152-3p mimic led to much higher levels of autophagy than DNMT1 overexpression alone (Fig. 3D). Western blot analysis results revealed a decrease in Beclin-1 protein expression and the LC3-II/LC3-I ratio but increases in DNMT1 and P62 protein expression in DOX-exposed H9c2 cells. DNMT1 overexpression decreased Beclin-1 protein expression and the LC3-II/LC3-I ratio while elevating DNMT1 and P62 protein expression. miR-152-3p overexpression caused the opposite trend. Simultaneous overexpression of DNMT1 and miR-152-3p increased Beclin-1 protein expression and the LC3-II/LC3-I ratio and reduced DNMT1 and P62 protein expression (Fig. 3E). These data supported that DNMT1 could attenuate mitophagy in H9c2 cells by limiting miR-152-3p.
DNMT1 upregulates the expression of ETS1 by inhibiting miR-152-3p
Intersection analysis of the first 100 target genes of miR-152-3p predicted by the miRDB and starBase databases using the bioinformatics database revealed five genes in the intersection: ETS1, AKAP1, FXR1, WDR47 and SOS2 (Fig. 4A). Published literature highlights the involvement of ETS1 in the progression of myocardial fibrosis-related disease, and inhibition of ETS-1 may represent a potential therapeutic strategy for the treatment of heart failure secondary to chronic hypertension12. Therefore, we chose to focus on ETS1 in subsequent experiments.
A Venn diagram analysis of the first 100 target genes of miR-152-3p predicted using the miRDB and starBase databases. B miR-152-3p binding sites in the 3′UTR of ETS1 mRNA were predicted using the TargetScan website. C Expression of ETS1 in heart tissues from control rats and DOX-treated rats determined by RT-qPCR. D Pearson’s correlation analysis of the relationship between ETS1 and miR-152-3p. E Pearson’s correlation analysis of the relationship between ETS1 and DNMT1. F Binding of miR-152-3p to ETS1 was verified by dual-luciferase reporter assay. G RT-qPCR of miR-152-3p expression and western blot analysis of ETS1 protein in H9c2 cells transfected with miR-152-3p mimic or miR-152-3p inhibitor. H DNMT1 overexpression and knockdown efficiency in H9c2 cells validated by western blot analysis and RT-qPCR. I Western blot analysis of ETS1 and DNMT1 proteins in H9c2 cells transfected with oe-DNMT1 or sh-DNMT1. J Western blot analysis of ETS1 and DNMT1 proteins in H9c2 cells transfected with oe-DNMT1, oe-DNMT1 + miR-152-3p mimic, sh-DNMT1 or sh-DNMT1 + miR-152-3p inhibitor. *p < 0.05, compared to control rats, cells transfected with mimic-NC, oe-NC, oe-NC + mimic NC or sh-NC + inhibitor NC; #p < 0.05, compared to cells transfected with inhibitor-NC, sh-NC, or oe-DNMT1 + mimic NC; &p < 0.05, compared to cells transfected with sh-DNMT1 + inhibitor NC. n = 10 rats in each group. The cell experiments were independently conducted three times.
The TargetScan website predicted the presence of binding sites between EST1 and miR-152-3p (Fig. 4B). More importantly, expression of ETS1 was upregulated in heart tissues from DOX-treated rats (Fig. 4C). Pearson’s correlation analysis revealed a negative correlation between ETS1 and miR-152-3p (Fig. 4D) and a positive correlation between ETS1 and DNMT1 in heart tissues from DOX-treated rats (Fig. 4E). A dual-luciferase reporter assay further confirmed that transfection with the miR-152-3p mimic significantly reduced luciferase activity of the ETS1-3′UTR-WT but did not affect that of the ETS1-3′UTR-MUT (Fig. 4F). Moreover, protein levels of ETS1 were promoted in response to miR-152-3p mimic transfection but were suppressed following transfection with miR-152-3p inhibitor (Fig. 4G). RT-qPCR and western blot data validated the transfection efficiency of oe-DNMT1 and sh-DNMT1 in myoblasts, and the sh-DNMT12 sequence, which had superior efficiency, was selected for subsequent analysis (Fig. 4H). The western blot results showed that DNMT1 overexpression increased ETS1 protein expression, while DNMT1 silencing decreased ETS1 protein expression in H9c2 cells (Fig. 4I). To further determine whether DNMT1 upregulation of ETS1 expression is miR-152-3p-dependent, we overexpressed or knocked down the expression of DNMT1 and/or miR-152-3p in myoblasts. The western blot results suggested that oe-DNMT1 promoted DNMT1 and ETS1 expression, while an additional miR-152-3p mimic abolished this promotion of ETS1 expression without altering DNMT1 expression (Fig. 4J). Therefore, DNMT1 could promote the expression of ETS1 by suppressing expression of miR-152-3p.
ETS1 suppresses mitophagy in H9c2 cells by promoting the expression of RhoH
After differential analysis of the GSE84796 dataset retrieved from the GEO database, we selected the top 20 genes for subsequent analysis (Fig. 5A). At the same time, a previous study highlighted the key role of the Rho kinase pathway in the pathogenesis of heart failure26, but its specific mechanism needs further exploration. Therefore, we used RhoH as a target for further study. The STRING database predicted an interaction between RhoH and ETS1 (Fig. 5B). Meanwhile, the ChIPbase database showed a significant positive correlation between RhoH and ETS1 in myocardial tissues (Fig. 5C), suggesting that ETS1 may act as a transcription factor to promote RhoH transcription by binding to the promoter region of RhoH.
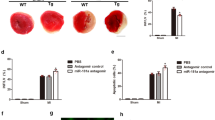
A A heatmap of differentially expressed genes related to heart failure obtained from the GSE84796 dataset. The x-axis represents the sample number, and the y-axis represents the gene name. B Interaction between RhoH and ETS1 analyzed using the STRING database. C Correlation between RhoH and ETS1 in myocardial tissues analyzed using the ChIPbase database. D Expression of RhoH in heart tissues from normal rats and DOX-treated rats determined by RT-qPCR. E Pearson’s correlation analysis of the relationship between ETS1 expression and RhoH expression. F western blot analysis of EST1 and RhoH proteins in H9c2 cells with ETS1 overexpression. G RhoH knockdown efficiency in H9c2 cells was confirmed by RT-qPCR and western blot analysis. H Viability of H9c2 cells treated with oe-ETS1, sh-RhoH or both measured by MTT assay. I LDH release in H9c2 cell culture solution in the presence of oe-ETS1, sh-RhoH or both. J MDC-positive H9c2 cells treated with oe-ETS1, sh-RhoH or both, scale bar = 25 μm. K Protein expression of ETS1, RhoH, P62, Beclin-1, LC3-II, and LC3-I in H9c2 cells treated with oe-ETS1, sh-RhoH or both determined by western blot analysis. *p < 0.05, compared to control rats, cells treated with oe-NC, sh-NC, or oe-NC + sh-NC + DOX; #p < 0.05, compared to cells treated with oe-ETS1 + sh-NC + DOX. n = 10 rats in each group. The cell experiments were independently conducted three times.
Elevation of RhoH was observed in heart tissues of DOX-treated rats (Fig. 5D); moreover, it was positively correlated with ETS1 expression (Fig. 5E). Western blot analysis further demonstrated that ETS1 overexpression promoted the expression of RhoH (Fig. 5F). H9c2 cells were transfected with sh-RhoH, the efficiency of which was confirmed by RT-qPCR and western blot analysis. sh-RHOH2 showed superior efficiency and was selected for further experiments (Fig. 5G, Supplemental Fig. 1C). ETS1 overexpression led to a decrease in cell viability and an increase in the LDH release rate, aggravating cell injury, while depletion of RhoH resulted in the opposite results. Combined transfection with oe-ETS1 and sh-RhoH repressed cell injury (Fig. 5H, I). Moreover, ETS1 upregulation suppressed autophagy, as reflected by decreased green fluorescence intensity and MDC-positive cells, while RhoH silencing promoted DOX-induced autophagy, as reflected by elevated green fluorescence intensity and MDC-positive cells. However, dual transfection with oe-ETS1 and sh-RhoH enhanced autophagy compared to oe-ETS1 alone (Fig. 5J). Furthermore, oe-ETS1 treatment suppressed mitophagy, while further RhoH depletion abrogated this trend. Additionally, increased mitophagy was observed in the presence of oe-ETS1 + sh-RhoH compared to oe-ETS1 alone (Fig. 5K). These data suggested that ETS1 suppressed mitophagy in myoblasts by promoting the expression of RhoH.
DNMT1 reduces the mitophagy in H9c2 cells through the miR-152-3p/ETS1/RhoH axis
The aforementioned results encouraged us to speculate that DNMT1 may repress mitophagy in myoblasts via the miR-152-3p/ETS1/RhoH axis. To test this hypothesis, we conducted relevant assays in H9c2 cells. RT-qPCR results revealed increased DNMT1 and decreased miR-152-3p expression in cells with oe-DNMT1, while DNMT1 and miR-152-3p expression exhibited no changes in the presence of oe-DNMT1 + sh-RhoH (Fig. 6A). In addition, western blot analysis showed that oe-DNMT1 elevated the expression of RhoH, DNMT1 and ETS1, but RhoH expression was reduced following further sh-RhoH treatment (Fig. 6B, Supplemental Fig. 1D). The results of MTT and LDH assays showed that DNMT1 overexpression reduced cell viability and increased the LDH release rate, inducing cell injury, while RhoH silencing or combination with DNMT1 overexpression attenuated cell injury (Fig. 6C, D). DNMT1-overexpressing cells displayed suppressed autophagy, and in contrast, RhoH silencing or combination with DNMT1 overexpression negated this trend (Fig. 6E, F). In addition, DNMT1 overexpression repressed mitophagy, which was neutralized by RhoH knockdown or combination with DNMT1 overexpression (Fig. 6G). Furthermore, H9c2 cells with parkin-KO or ATG7-KO exhibited higher DNMT1, ETS1 and RhoH expression and lower miR-152-3p expression along with reduced autophagy compared to control cells (Fig. 6H, I), indicating that DNMT1 promotes myoblast injury by regulating the miR-152-3p/ETS1/RhoH axis through the suppression of mitophagy. Overall, these results indicated that DNMT1 inhibited mitophagy in myoblasts through the miR-152-3p/ETS1/RhoH axis.
A Expression of DNMT1 and miR-152-3p in H9c2 cells treated with oe-DNMT1 or combined with sh-RhoH determined by RT-qPCR. B Western blot analysis of DNMT1, ETS1 and RhoH proteins in H9c2 cells treated with oe-DNMT1 or combined with sh-RhoH. C Viability of H9c2 cells treated with oe-DNMT1 or combined with sh-RhoH measured by MTT assay. D LDH release in H9c2 cell culture solution in the presence of oe-DNMT1 alone or in combination with sh-RhoH. E MDC-positive H9c2 cells treated with oe-DNMT1 alone or in combination with sh-RhoH. Scale bar = 25 μm. F Colocation ratio. G Western blot analysis of Beclin-1, P62, and LC3-II/LC3-I in H9c2 cells treated with oe-DNMT1 or combined with sh-RhoH. H Western blot analysis of DNMT1, ETS1, RhoH, ATG7, parkin, P62 and LC3-II/LC3-I in H9c2 cells treated with oe-DNMT1 alone or in combination with sh-RhoH. I Expression of miR-152-3p in H9c2 cells treated with oe-DNMT1 or combined with sh-RhoH determined by RT-qPCR. *p < 0.05, compared to control cells or cells treated with oe-NC + sh-NC; #p < 0.05, compared to cells treated with oe-DNMT1 + sh-NC. The cell experiments were independently conducted three times.
Depletion of DNMT1 protects rats from DOX-induced heart failure through the miR-152-3p/ETS1/RhoH axis
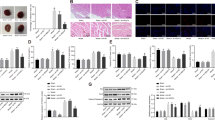
Our data demonstrated that DNMT1 overexpression suppressed mitophagy in myoblasts by regulating the miR-152-3p/ETS1/RhoH axis in vitro. To further confirm our findings in vivo, we established DOX-induced rat heart failure models. DOX-exposed rats displayed reduced LVEF and LVFS, as well as elevated LVDD and LVDS. Importantly, the echocardiography parameters of DOX-exposed rats were recovered by DNMT1 silencing to a comparable level to that of control rats. Moreover, this protective effect achieved by DNMT1 silencing was diminished by further depletion of RhoH, as reflected by reduced LVEF and LVFS, as well as increased LVDD and LVSD (Supplemental Table 3). H&E staining revealed that tissues from DOX-treated rats were severely damaged; however, in the sh-DNMT1 + LV-NC group, tissue damage was alleviated, as indicated by the normal arrangement of the muscle fibers and reduced inflammatory cell infiltration. Moreover, upon cotreatment with sh-DNMT1 and LV-RhoH, cardiomyopathy-like changes were observed in myocardial tissues relative to sh-DNMT1 treatment alone (Fig. 7A). In-gel ATPase activity and JC-1 assays demonstrated that DOX treatment reduced ATPase activity and mitochondrial membrane potential, while depletion of DNMT1 recovered the activity of mitochondrial ATPase and mitochondrial membrane potential. In addition, combined treatment with sh-DNMT1 and LV-RhoH led to reduced ATPase activity and mitochondrial membrane potential compared to treatment with sh-DNMT1 alone (Fig. 7B, C). Furthermore, RT-qPCR and western blot were performed to assess miR-152-3p DNMT1, ETS1, RHOH, and the autophagy-related factors Beclin-1, LC3-II, LC3-I and P62 in the myocardial tissue of each group of rats. In addition, expression of DNMT1, ETS1, RhoH and P62 was upregulated, while that of miR-152-3p and Beclin-1 along with the LC3-II/LC3-I ratio was reduced in the heart tissues of DOX-treated rats, and treatment with sh-DNMT1 resulted in the opposite findings. Combined treatment with sh-DNMT1 and LV-RhoH induced higher RhoH and P62 expression but lower Beclin-1 expression and LC3-II/LC3-I ratio compared to sh-DNMT1 alone (Fig. 7D, E). These data suggested that the protective effect of DNMT1 depletion on heart failure was correlated with the miR-152-3p/ETS1/RhoH axis.
A Representative images of H&E staining in heart tissues from rats treated with sh-DNMT1 or combined with RhoH. Black arrow indicates neutrophil infiltration. Scale bar = 50 μm. B ATPase activity measurement in heart tissues from rats treated with sh-DNMT1 or combined with RhoH. C Mitochondrial membrane potential in rats treated with sh-DNMT1 or combined with RhoH measured by JC-1 assay. D Expression of DNMT1, miR-152-3p, EST1 and RhoH in heart tissues from rats treated with sh-DNMT1 or combined with RhoH determined by RT-qPCR. E Western blot analysis of DNMT1, ETS1, RhoH, Beclin-1, and P62 proteins and LC3-II/LC3-I ratio in heart tissues from rats treated with sh-DNMT1 or combined with RhoH. *p < 0.05, compared to control rats or DOX-induced rats treated with sh-NC + LV-NC; #p < 0.05, compared to DOX-induced rats treated with sh-NC + LV-NC; &p < 0.05, compared to DOX-induced rats treated with sh-DNMT1 + LV-NC. n = 10 rats in each group.
Discussion
Heart failure, a major public health burden, affects ~2% of the adult population worldwide. According to a recent study, heart failure-related mortality has not meaningfully improved since 2009. Moreover, the estimated mean cost for hospitalization with primary heart failure will be 11 billion27. Therefore, there is an urgent need for a better understanding of the pathophysiology and pathogenesis of heart failure. DNA methylation has been reported to show great potential for regulating multiple types of biological processes essential for cardiac function28. In the current study, we focused on the role of one of the DNA methyltransferase family members, DNMT1, in the pathogenesis of heart failure. We concluded that DNMT1 could inhibit mitophagy in H9c2 cells and facilitates the ensuing occurrence of heart failure through regulation of the miR-152-3p/ETS1/RhoH axis.
DNMT1 is the most abundant and major maintenance methyltransferase among DNMT family members and primarily methylates hemimethylated DNA29. DNMT1 has been studied in multiple types of diseases, including cancer30,31, neurological disorders32,33, and genetic diseases34. Recently, a study showed that cardiomyocytes with specific depletion of DNMT1 protected rats from DOX-induced heart failure7. In addition, DNMT1 is highly expressed in heart failure rats and hypertrophy cell models, and this high expression suppresses SHP-1 expression, leading to the development of cardiomyocyte hypertrophy-induced heart failure35. In this study, we determined the role of DNMT1 in the initiation and progression of heart failure and defined the underlying mechanism. We induced heart failure using DOX in vivo and in vitro, and we observed that protein levels of DNMT1 were upregulated in DOX-treated rats and H9c2 cells.
We also observed that DOX treatment resulted in the downregulation of miR-152-3p and that expression of DNMT1 and miR-152-3p was negatively correlated in DOX-treated rats. In alignment with previous literature, we showed that DNMT1 inhibited the expression of miR-152-3p by methylating its promoter region9. miR-152-3p has demonstrated cardioprotective roles in heart failure due to its inhibitory effect on the apoptosis of H9C2 cells24. We also found that DNMT1 overexpression reduced the expression of miR-152-3p, while reconstitution of miR-152-3p in DOX-treated H9c2 cells restored cell viability. Because dysregulated mitophagy can reduce the viability of myocardial cells18, we speculated that miR-152-3p protected H9c2 cells by enhancing mitophagy. Therefore, we investigated the effect of miR-152-3p in DOX-treated myocardial cells. We observed that miR-152-3p reconstitution significantly enhanced autophagy and mitophagy in H9c2 cells.
We also concluded that miR-152-3p targeted ETS1 and downregulated the expression of ETS1 and that ETS1-regulated RhoH in myoblasts. The significance of miRNAs in heart failure through modulation of mRNA has been documented36. Partly in line with our findings, a recent study highlighted that mice with miR-152 overexpression exhibited reduced DOX-related cardiac injury through modulation of Nrf237. ETS1 is a key regulator of vascular homoeostasis and is critical for cardiac neural crest and heart development38,39. Deletion of ETS1 has been reported to attenuate angiotensin-II-induced cardiac fibrosis by repressing endothelial-to-mesenchymal transition, which represents a potential therapeutic strategy for the treatment of heart failure secondary to chronic hypertension12. Overexpression of ETS1 has been shown to upregulate the promoter activity and mRNA and protein expression of RhoC, a member of the Rho subfamily of the Ras superfamily of homologous genes40. Interestingly, Rho-kinase inhibition is cardioprotective against diclofenac toxicity, as it can prevent diclofenac-induced increases in myocardial oxidative damage markers41. In addition, downregulation of Rho/ROCK confers protection to cardiomyocytes against heart failure, of which Rho A expression is significantly increased in the myocardial tissues of rats with heart failure42. Rho-kinase pathway inhibition ameliorates diastolic heart failure, as evidenced by decreased myocardial stiffness, cardiomyocyte hypertrophy, cardiac fibrosis and superoxide production, represents a promising therapeutic target for this disorder26. Therefore, it is reasonable to conclude that miR-152-3p may target ETS1 and downregulate the expression of ETS1 as well as ETS1-mediated RhoH in H9c2 cells. It is also possible that in DOX-treated H9c2 cells, downregulated miR-152-3p resulted in a reduction in cell mitophagy and viability by de-suppressing ETS1/RhoH. To test this hypothesis, we modulated the expression of miR-152-3p, ETS1 and RhoH in H9c2 cells. We then evaluated H9c2 cell viability, autophagy and mitophagy in cells treated with or without DOX. Our data also demonstrated that DNMT1 overexpression-mediated miR-152-3p reduction resulted in upregulated ETS1 and RhoH in H9c2 cells. Moreover, we showed that in DOX-treated H9c2 cells, depletion of RhoH was enhanced, while ETS1 overexpression suppressed mitophagy and cell viability.
These findings were also validated in a rat model of heart failure in which DNMT1 depletion significantly improved cardiac function, as reflected by improved LVEF and LVFS, as well as alleviated LVSD and LVDD, consistent with previous studies7. Moreover, DNMT1 silencing improved autophagy in vivo, which has been demonstrated by others43. These effects were compromised by further RhoH overexpression. These findings indicate that miR-152-3p/ETS1/RhoH may function as an axis that regulates autophagy and mitophagy and ultimately regulates H9c2 cell viability during heart failure. Moreover, as our data showed, during heart failure, DNMT1 was upregulated, which, in turn, suppressed the expression of miR-152-3p.
Our study bridges the gap between DNMT1-regulated miR-152-3p and ETS1-regulated RhoH in the pathogenesis of heart failure. Our data demonstrate that, mechanistically, during heart failure, upregulated DNMT1 inhibits the expression of miR-152-3p, which further results in upregulation of ETS1 and RhoH. Functionally, we showed that heart failure-mediated upregulation of DNMT1 suppressed mitophagy in H9c2 cells and consequently reduced the viability of those cells (Fig. 8). Our study identified the role of DNMT1 in the pathogenesis of heart failure and the underlying molecular mechanisms. This study sheds light on the development of therapeutic strategies targeting DNMT1 in heart failure. However, additional studies are required to validate the findings in our study due to the lack of supporting literature regarding the correlation between ETS1 and RhoH with respect to the role of RhoH in heart failure. Furthermore, the mechanism by which DNMT1 regulates mitophagy through the miR-152-3p/ETS1/RHOH signaling axis and affects the development of heart failure may be related to chemotherapy-induced CM, but the role of this mechanism in other myocardial diseases remains to be explored.
Data availability
All data generated or analyzed during this study are included in this article [and/or] its supplemental material files. Further enquiries can be directed to the corresponding author.
References
Ziaeian, B. & Fonarow, G. C. Epidemiology and aetiology of heart failure. Nat. Rev. Cardiol. 13, 368–378 (2016).
Mazurek, J. A. & Jessup, M. Understanding Heart Failure. Heart Fail. Clin. 13, 1–19 (2017).
Xu, R., Sun, Y., Chen, Z., Yao, Y. & Ma, G. Hypoxic Preconditioning Inhibits Hypoxia-induced Apoptosis of Cardiac Progenitor Cells via the PI3K/Akt-DNMT1-p53 Pathway. Sci. Rep. 6, 30922 (2016).
Jorgensen, B. G. et al. DNA methylation, through DNMT1, has an essential role in the development of gastrointestinal smooth muscle cells and disease. Cell Death Dis. 9, 474 (2018).
Vukic, M. & Daxinger, L. DNA methylation in disease: Immunodeficiency, Centromeric instability, Facial anomalies syndrome. Essays Biochem. 63, 773–783 (2019).
Papait, R., Serio, S. & Condorelli, G. Role of the Epigenome in Heart Failure. Physiol. Rev. 100, 1753–1777 (2020).
Wu, T. T. et al. Myocardial tissue-specific Dnmt1 knockout in rats protects against pathological injury induced by Adriamycin. Lab. Investig. 100, 974–985 (2020).
Chaturvedi, P. et al. Differential regulation of DNA methylation versus histone acetylation in cardiomyocytes during HHcy in vitro and in vivo: an epigenetic mechanism. Physiol. Genom. 46, 245–255 (2014).
Ramalho-Carvalho, J. et al. A multiplatform approach identifies miR-152-3p as a common epigenetically regulated onco-suppressor in prostate cancer targeting TMEM97. Clin. Epigenet. 10, 40 (2018).
Lu, Z. W. et al. MiR-152 functioning as a tumor suppressor that interacts with DNMT1 in nasopharyngeal carcinoma. Onco. Targets Ther. 11, 1733–1741 (2018).
Zhang J., et al. Overexpression of Exosomal Cardioprotective miRNAs Mitigates Hypoxia-Induced H9c2 Cells Apoptosis. Int. J. Mol. Sci. 18, 711 (2017).
Xu, L. et al. Endothelial-specific deletion of Ets-1 attenuates Angiotensin II-induced cardiac fibrosis via suppression of endothelial-to-mesenchymal transition. BMB Rep. 52, 595–600 (2019).
Troeger, A. & Williams, D. A. Hematopoietic-specific Rho GTPases Rac2 and RhoH and human blood disorders. Exp. Cell Res. 319, 2375–2383 (2013).
Yan, Y., Xiang, C., Yang, Z., Miao, D. & Zhang, D. Rho Kinase Inhibition by Fasudil Attenuates Adriamycin-Induced Chronic Heart Injury. Cardiovasc. Toxicol. 20, 351–360 (2020).
Santos, G. L., Hartmann, S., Zimmermann, W. H., Ridley, A. & Lutz, S. Inhibition of Rho-associated kinases suppresses cardiac myofibroblast function in engineered connective and heart muscle tissues. J. Mol. Cell Cardiol. 134, 13–28 (2019).
Ding, R., Han, J., Zhao, D., Hu, Z. & Ma, X. Pretreatment with Rho-kinase inhibitor ameliorates lethal endotoxemia-induced liver injury by improving mitochondrial function. Int. Immunopharmacol. 40, 125–130 (2016).
Zhou, H., He, L., Xu, G. & Chen, L. Mitophagy in cardiovascular disease. Clin. Chim. Acta 507, 210–218 (2020).
Moyzis, A. G., Sadoshima, J. & Gustafsson, A. B. Mending a broken heart: the role of mitophagy in cardioprotection. Am. J. Physiol. Heart Circ. Physiol. 308, H183–H192 (2015).
Lu, L. et al. Adriamycin-induced autophagic cardiomyocyte death plays a pathogenic role in a rat model of heart failure. Int. J. Cardiol. 134, 82–90 (2009).
Wen, J. et al. Salsolinol Attenuates Doxorubicin-Induced Chronic Heart Failure in Rats and Improves Mitochondrial Function in H9c2 Cardiomyocytes. Front. Pharmacol. 10, 1135 (2019).
Hoshino, A. et al. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat. Commun. 4, 2308 (2013).
Ma, L. Q. et al. Sweroside Alleviated Aconitine-Induced Cardiac Toxicity in H9c2 Cardiomyoblast Cell Line. Front. Pharmacol. 9, 1138 (2018).
Xu, Q. et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 5, 3–13 (2013).
Zhang, Z. et al. Tanshinone IIA inhibits apoptosis in the myocardium by inducing microRNA-152-3p expression and thereby downregulating PTEN. Am. J. Transl. Res. 8, 3124–3132 (2016).
Momparler, R. L., Cote, S., Momparler, L. F. & Idaghdour, Y. Inhibition of DNA and Histone Methylation by 5-Aza-2’-Deoxycytidine (Decitabine) and 3-Deazaneplanocin-A on Antineoplastic Action and Gene Expression in Myeloid Leukemic Cells. Front. Oncol. 7, 19 (2017).
Fukui, S. et al. Long-term inhibition of Rho-kinase ameliorates diastolic heart failure in hypertensive rats. J. Cardiovasc. Pharmacol. 51, 317–326 (2008).
Jackson, S. L. et al. National Burden of Heart Failure Events in the United States, 2006 to 2014. Circ. Heart Fail. 11, e004873 (2018).
Pepin, M. E. et al. DNA methylation reprograms cardiac metabolic gene expression in end-stage human heart failure. Am. J. Physiol. Heart Circ. Physiol. 317, H674–H684 (2019).
Fernandes, G. F. S. et al. Epigenetic Regulatory Mechanisms Induced by Resveratrol. Nutrients 9, 1201 (2017).
Wang, P. et al. Kindlin-2 interacts with and stabilizes DNMT1 to promote breast cancer development. Int. J. Biochem. Cell Biol. 105, 41–51 (2018).
Du, W. W. et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 37, 5829–5842 (2018).
Higuchi, F. et al. State-dependent changes in the expression of DNA methyltransferases in mood disorder patients. J. Psychiatr. Res. 45, 1295–1300 (2011).
Fuso, A., Nicolia, V., Cavallaro, R. A. & Scarpa, S. DNA methylase and demethylase activities are modulated by one-carbon metabolism in Alzheimer’s disease models. J. Nutr. Biochem. 22, 242–251 (2011).
Sacconi, S. et al. Patients with a phenotype consistent with facioscapulohumeral muscular dystrophy display genetic and epigenetic heterogeneity. J. Med. Genet. 49, 41–46 (2012).
Wang Y. Y., et al. Histone deacetylase 3 suppresses the expression of SHP-1 via deacetylation of DNMT1 to promote heart failure. Life Sci. 119552, https://doi.org/10.1016/j.lfs.2021.119552 (2021).
Vegter, E. L., van der Meer, P., de Windt, L. J., Pinto, Y. M. & Voors, A. A. MicroRNAs in heart failure: from biomarker to target for therapy. Eur. J. Heart Fail. 18, 457–468 (2016).
Zhang, W. B., Lai, X. & Guo, X. F. Activation of Nrf2 by miR-152 Inhibits Doxorubicin-Induced Cardiotoxicity via Attenuation of Oxidative Stress, Inflammation, and Apoptosis. Oxid. Med. Cell Longev. 2021, 8860883 (2021).
Sindi, H. A. et al. Therapeutic potential of KLF2-induced exosomal microRNAs in pulmonary hypertension. Nat. Commun. 11, 1185 (2020).
Gandhi, S., Ezin, M. & Bronner, M. E. Reprogramming Axial Level Identity to Rescue Neural-Crest-Related Congenital Heart Defects. Dev. Cell 53, 300–315 e304 (2020).
Qin, D. et al. HBx and HBs regulate RhoC expression by upregulating transcription factor Ets-1. Arch. Virol. 158, 1773–1781 (2013).
Arafa, M. H., Mohammad, N. S. & Atteia, H. H. Rho-Kinase inhibitors ameliorate diclofenac-induced cardiotoxicity in chloroquine-treated adjuvant arthritic rats. Life Sci. 254, 117605 (2020).
Yang, T. et al. Ginsenoside Rb1 inhibits autophagy through regulation of Rho/ROCK and PI3K/mTOR pathways in a pressure-overload heart failure rat model. J. Pharm. Pharmacol. 70, 830–838 (2018).
Yang, A. et al. Homocysteine activates autophagy by inhibition of CFTR expression via interaction between DNA methylation and H3K27me3 in mouse liver. Cell Death Dis. 9, 169 (2018).
Author information
Authors and Affiliations
Contributions
Z.J.D., J.Q.Y., and X.Y.G. conceived and designed research. J.Q.Y., N.X., and Y.H. performed experiments. N.X., Y.H., and X.W. analyzed data. C.Z.C. and K.C. interpreted results of experiments. Z.J.D. prepared figures. Z.J.D., N.X., Y.H., and X.W. drafted paper. J.Q.Y., C.Z.C., K.C., and X.Y.G. edited and revised paper. All authors approved final version of paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The current study was approved by the Animal Ethics Committee of the Third Hospital of Hebei Medical University. We attempted to reduce the suffering of the animal used during the experiments.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Deng, Z., Yao, J., Xiao, N. et al. DNA methyltransferase 1 (DNMT1) suppresses mitophagy and aggravates heart failure via the microRNA-152-3p/ETS1/RhoH axis. Lab Invest 102, 782–793 (2022). https://doi.org/10.1038/s41374-022-00740-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-022-00740-8
This article is cited by
-
SGLT2 inhibitors: role in protective reprogramming of cardiac nutrient transport and metabolism
Nature Reviews Cardiology (2023)