Abstract
Glioma stem cells (GSCs) are self-renewing tumor cells with multi-lineage differentiation potential and the capacity of construct glioblastoma (GBM) heterogenicity. Mitochondrial morphology is associated with the metabolic plasticity of GBM cells. Previous studies have revealed distinct mitochondrial morphologies and metabolic phenotypes between GSCs and non-stem tumor cells (NSTCs), whereas the molecules regulating mitochondrial dynamics in GBM cells are largely unknown. Herein, we report that carnitine palmitoyltransferase 1A (CPT1A) is preferentially expressed in NSTCs, and governs mitochondrial dynamics and GSC differentiation. Expressions of CPT1A and GSC marker CD133 were mutually exclusive in human GBMs. Overexpression of CPT1A inhibited GSC self-renewal but promoted mitochondrial fusion. In contrast, disruption of CPT1A in NSTCs promoted mitochondrial fission and reprogrammed NSTCs toward GSC feature. Mechanistically, CPT1A overexpression increased the phosphorylation of dynamin-related protein 1 at Ser-637 to promote mitochondrial fusion. In vivo, CPT1A overexpression decreased the percentage of GSCs, impaired GSC-derived xenograft growth and prolonged tumor-bearing mice survival. Our work identified CPT1A as a critical regulator of mitochondrial dynamics and GSC differentiation, indicating that CPT1A could be developed as a molecular target for GBM cell-differentiation strategy.
Similar content being viewed by others
Introduction
Glioblastoma (GBM) is the most common and lethal primary brain tumor. Therapeutic regimens for GBM include maximized surgical resection, radiation, and chemotherapy1. However, the median survival of GBM patients is <16 months2. GBM stem cells (GSCs) are a subset of glioma cells with self-renewal ability, multi-differentiation potential and tumor-initiating capacity. Previous works have demonstrated that GSCs are privileged to drive tumorigenesis, tumor invasion and chemo-/radio-resistance3. GSCs also actively interact with stromal cells in GBM microenvironment to promote tumor angiogenesis and immune evasion4. The evidence showing GSCs as a hub for GBM initiation and progression suggests that strategies targeting GSC survival or reprogramming GSCs toward a differentiation state are critical for improving GBM patient outcome. It has been reported that cancer stem cells (CSCs) are potentiated by increased glycolysis than non-stem tumor cells (NSTCs) in malignant tumors such as GBMs, breast cancers, and lung cancers5,6. CSCs also rely on fatty acid oxidation for self-renewal and chemoresistance7. Identification of the special metabolic characteristics of CSCs relative to NSTCs could be manipulated for the development of a therapeutic strategy targeting CSC populations.
Mitochondria are the central metabolic organelles that undergo dynamic morphology changes (fusion and fission) under different physiological or pathological conditions. Mitochondrial fission leads to fragmented mitochondria, while mitochondrial fusion is defined as a state that more than two mitochondria merge into one mitochondrial network8. Mitochondrial fission is mainly regulated by dynamin-related protein 1 (DRP1), which can be recruited to the outer membrane of mitochondria, to form a ring-like structure and to promote mitochondrial fission9. The GTPase inhibitor of DRP1 such as mdivi-1 promotes DRP1 phosphorylation at Ser-637 but inhibits DRP1 phosphorylation at Ser-616, thus inhibiting mitochondrial fission10. Mitochondrial fusion is mainly mediated by dynamin GTPases Mitofusin (MFN) and Optic atrophy 1 (OPA1). MFN is in charge of the fusion of mitochondrial outer membrane, while OPA1 regulates the fusion of mitochondrial inner membrane11. Previous studies have shown that GSC mitochondria were different from that of NSTCs. GSCs have a low mitochondrial number, reduced mitochondrial DNA copy number and lower mitochondrial reactive oxygen species (ROS) level than NSTCs7. GSC differentiation requires mitochondrial DNA replication. Mitochondrial DNA depletion inhibits GSC differentiation and increases the expressions of pluripotency genes in GSCs12. Reduced ROS level is also required for the maintenance of CSC stemness. Increased mitochondrial ROS facilitates CSC differentiation5,6. Mitochondrial dynamics varies between GSCs and NSTCs. In GSCs, mitochondria show a fission state, while in NSTCs, mitochondria display a fusion state. Inhibition of mitochondrial fission promotes GSC differentiation and inhibits the growth of GSC-derived xenografts13. However, the molecules governing mitochondrial dynamics in GSCs and NSTCs remain largely unknown.
Carnitine palmitoyltransferase 1A (CPT1A), a rate-limiting enzyme regulating fatty acid transport and oxidation, is located on the outer membrane of mitochondria14. It has been suggested that the fundamental unit of CPT1A interacts with long-chain acyl-coenzyme A synthetase and voltage-dependent anion channel on mitochondrial outer membrane to create a hexamer channel for the trafficking activated fatty acids into mitochondria, thus facilitating fatty acid utilization and maximizing ATP production15. CPT1A is widely expressed in multiple organs and tissues14. Dysfunction of CPT1A is associated with impaired mitochondrial functions and contributes to metabolic diseases or disorders including hepatic steatosis, Alzheimer’s disease, obesity, and atherosclerosis14. CPT1A overexpression augments ROS in the mitochondria and supports prostate cancer progression16. Targeting CPT1A makes nasopharyngeal carcinoma more sensitive to radiation therapy15. As the expression of CPT1A in human GBMs and its function in GBM biology remain largely unknown, we investigated the role of CPT1A in regulating GBM cell mitochondrial dynamics and GSC differentiation.
Materials and methods
GBM specimens
Human primary GBM specimens were collected from the Biobank of Southwest Hospital (Chongqing, China), approved by the Ethics Committees of Southwest Hospital. Fresh GBM tissues were dissociated into single cells for primary cell culture or made into frozen sections for immunofluorescent staining.
Cell isolation and culturing
GBM cells were dissociated from primary GBM surgical specimens by using papain disassociation system (Worthington Biochemical, Lakewood, NJ, USA), followed by fluorescence-activated cell sorting (FACS) of GSCs and NSTCs according to the protocols mentioned in previous studies17. In brief, the isolated GBM cells were stained with PE-conjugated anti-CD133 antibody (Miltenyi, Bergisch Gladbach, Germany) along with FITC-conjugated anti-CD15 antibody (BD Biosciences, Heidelberg, Germany) at 4 °C for 30 min. FACS was applied to isolate CD133+/CD15+ GSCs and CD133−/CD15− NSTCs. The sorted cells were cultured in Neurobasal medium (Gibco, Waltham, MA, USA), supplemented with B27 supplement (Gibco), epidermal growth factor (PeproTech, Suzhou, China) and basic fibroblast growth factor (PeproTech). To validate the stemness of isolated GSCs, the GSC markers (CD133, SRY-box transcription factor 2 (SOX2), Oligodendrocyte Transcription Factor 2 (OLIG2) and CD15) were detected by immunoblotting, followed by functional assays including the serial tumor-sphere formation assay to detect the self-renewal potential of GSCs, and the in vivo limiting dilution assay to detect the tumor propagation4,17. For serum-induced GSC differentiation assay, GSCs were cultured in DMEM supplemented with 10% serum. To inhibit DRP1-mediated mitochondrial fission, NSTCs were treated with mdivi-1 at the concentration of 10 μM (Millipore, Beijing, China). All cells were cultured at 37 °C with 5% CO2/95% air and were confirmed as Mycoplasma and Acholeplasma free.
Immunofluorescent staining
Cells or 8 µm-thick tissue sections were fixed in 4% paraformaldehyde and stained using the following antibodies: TOM20 (Santa Cruz, Shanghai, China); SOX2 (Bethyl, Montgomery, TX, USA) and Ki67 (Abcam, Boston, MA, USA). Sections were incubated with primary antibodies at 4 °C for 15 h, followed by incubation of secondary antibodies (Invitrogen, Waltham, MA, USA) at room temperature for 1 h. The sections were then mounted with anti-fluorescence quencher containing DAPI (Beyotime Biotechnology, Shanghai, China). Images were acquired by Zeiss 900 confocal microscopy (Zeiss, Jena, Germany).
Lentiviral infection
GSCs were transfected with the pRRLSIN-cPPT-SFFV-MCS-SV40-puromycin lentiviral vector carrying CPT1A (GENE bio, Shanghai, China) to construct CPT1A-overexpressing GSCs. To disrupt CPT1A expression, NSTCs were transfected with the pRRLSIN-cPPT-U6-shRNA-SFFV-EGFP-SV40-puromycin lentiviral vector (GENE bio) expressing shRNAs targeting CPT1A (sh1-CPT1A, sh2-CPT1A). The shRNA sequences were listed as follows: sh1-CPT1A: 5′-TCCCTGAACACCGCAAATCTT-3′; sh2-CPT1A: 5′-AGCCATGAAGCTCTTAGACAA-3′. Targeting efficiency of lentiviral sh-CPT1A clones was confirmed by immunoblotting.
Immunoblot assay
Immunoblot assay was performed as previously described17. In brief, cells were collected and lysed using RIPA (Beyotime Biotechnology). Protein concentration was detected by using BCA protein concentration determination kit (Beyotime Biotechnology). Primary antibodies used in this study include: CPT1A (Proteintech, Rosemont, PA, USA); SOX2 (Bethyl); OLIG2 (Millipore); GFAP (Invitrogen); DRP1 (Abcam); phospho-DRP1(Ser-616) (Cell Signaling, Danvers, MA, USA); phospho-DRP1(Ser-637) (Cell Signaling); FIS1 (Abcam); MFF (Abcam); Mitofusion-1 (Abcam); Mitofusion-2 (Abcam); OPA1 (Abcam) and β-tubulin (Cell Signaling).
Quantitative real-time PCR
mRNA was isolated using the RNeasy kit (Fastagen, Hefei, China) and reversely transcribed into cDNA using PrimeScript RT reagent kit with gDNA Eraser (TAKARA, Shiga, Japan). Quantitative real-time PCR assay was performed using SYBR Green Master mix (TAKARA). The primers used in this study include: CPT1A-F:5′-TCCAGTTGGCTTATCGTGGTG-3′; CPT1A-R:5′-TCCAGAGTCCGATTGATTTTTGC-3′; TUBB3-F:5′-GGCCAAGGGTCACTACACG-3′; TUBB3-R:5′-GCAGTCGCAGTTTTCACACTC-3′.
Cell sphere formation assay
Cells were collected at logarithmic phase and 500 cells were added to 96-well plates with ten repeats in each group. Cells were cultured in Neurobasal medium (Gibco) at 37 °C with 5% CO2 for 7 days. On day 7, the number of tumor-spheres and sphere diameters were recorded under light microscope (Leica, Wetzlar, Germany).
Intracranial GSC-derived xenografts
GSC-derived intracranial xenografts were used to evaluate the effect of CPT1A on tumor growth. Six-week-old female non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice (Gempharmatech, Nanjing, China) were anesthetized followed by implantation of the indicated GSCs (2 × 104/per mouse). GSCs were injected into the right striatum adjacent to the corpus callosum as described previously17. To evaluate the growth of GSC-derived xenografts, mouse brains were scanned using magnetic resonance imaging (MRI) with T2 weighted imaging (Bruker, Billerica, MA, USA). Tumor volumes were evaluated by the program Analyze 5.0 (Analyze Direct, Overland Park, KS, USA). All animal experiments were approved by the Institutional Animal Care and Use Committee of Southwest Hospital, and were in accordance with the Guide for the Care and Use of Laboratory Animals.
Electron microscopy
Xenograft tumors were cut into 1 × 1 × 2 mm3 blocks and fixed in 2.5% glutaraldehyde in 0.1 M sodium phosphate overnight at 4 °C. After being washed with 0.1 M sodium phosphate buffer, tumor tissues were fixed by 1% osmium tetroxide for 1 h and washed with 0.1 M sodium phosphate buffer. Tissues were then dehydrated using 30–100% ethanol, wrapped with resin, and made for ultra-thin sections. Sections were observed by electron microscope and mitochondria were analyzed for at least 100 cells in each group (n = 6). Image J software was applied to measure mitochondria morphology.
Statistical analysis
All data were presented as mean ± standard deviation using GraphPad Prism 6.0 (San Diego, CA, USA). The differences between two groups were performed with the unpaired Student’s t test (two-tailed). One-way ANOVA followed by Dunnett’s multiple comparison test (GraphPad 6.0) was used to determine the significance among more than two groups. p value < 0.05 was considered statistically significant. Bivariate correlation analysis (Pearson’s r test) was used to analyze the correlation between two variables. Statistical differences were considered significant at *p < 0.05 and **p < 0.01.
Results
CPT1A is highly expressed in NSTCs
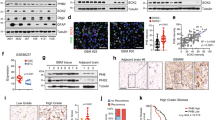
To determine the expression pattern of CPT1A in GSCs and NSTCs, we performed immunofluorescent staining of CPT1A and GSC marker CD133 in human GBM specimens. We found that CPT1A was preferentially expressed in CD133- NSTCs (Fig. 1a, b). CD133+ GSCs and CD133− NSTCs were then sorted from two human GBM specimens (GBM-1, -2) by flow cytometry. CPT1A was highly expressed in NSTCs compared with GSCs (Fig. 1c). The preferential expression of SOX2, a stem cell marker, confirmed the cell properties of isolated GSCs and NSTCs (Fig. 1d). To interrogate whether CPT1A expression was associated with GSC status, we detected CPT1A expression during GSC differentiation. Upon serum-induced differentiation, the expressions of GSC markers SOX2 and OLIG2 were gradually reduced while the astrocytic differentiation marker GFAP were increased (Fig. 1e). We also found a gradual increase of CPT1A during GSC differentiation (Fig. 1e), suggesting a positive correlation between CPT1A expression and GSC differentiation.
a Immunofluorescent staining of GSC marker CD133 (green) and CPT1A (red) in GBM tissues. Scale bars, 25 μm. Arrow indicates CD133+ cell, “#” indicates CPT1A+ cell. b Correlation analysis of the mean fluorescence intensity (MFI) of CPT1A and CD133 (n = 67). c Quantification of CPT1A mRNA in GSCs and NSTCs. d Immunoblot analysis of CPT1A and SOX2 in GSCs and NSTCs derived from GBM1 and GBM2. e Immunoblot analysis of CPT1A, GFAP, OLIG2, and SOX2 during GSC differentiation. Experiments in d, e were independently performed for at least three times. **p < 0.01.
CPT1A overexpression promotes GSC differentiation and impairs GSC self-renewal
To address whether CPT1A regulates GSC differentiation, we transduced CPT1A-expressing vector or control vector into GSCs, and found that CPT1A overexpression in GSCs inhibited the expression of GSC marker OLIG2 and SOX2, but increased the expression of astrocyte differentiation marker GFAP (Fig. 2a), suggesting that enforced CPT1A could promote GSC differentiation. In contrast, CPT1A disruption in NSTCs induced the expression of OLIG2 and SOX2, and inhibited GFAP expression (Fig. 2b), indicating that silencing CPT1A expression in NSTCs could reverse astrocyte differentiation state of GBM cells toward a GSC state. To determine the effect of CPT1A on regulating GSC self-renewal, we performed tumor-sphere formation assay and found that CPT1A overexpression significantly reduced the number and size of GSC tumor-spheres (Fig. 2c, d).
a Immunoblot analysis of CPT1A, GFAP, OLIG2, and SOX2 in NC or CPT1A-overexpression (CPT1A-OE) GSCs derived from GBM1 and GBM2. b Immunoblot analysis of CPT1A, GFAP, OLIG2, and SOX2 in NSTCs with or without CPT1A-knockdown (sh1-CPT1A and sh2-CPT1A). c Tumor-sphere formation assay of GSCs overexpressing CPT1A or not. d Quantification of tumor-sphere number and diameter of the GSCs overexpressing CPT1A or not. Experiments were independently performed for at least three times. **p < 0.01.
CPT1A overexpression promotes mitochondrial fusion in GSCs
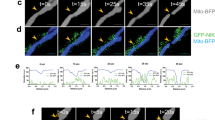
The role of CPT1A in transporting fatty acid components across the mitochondria prompted us to investigate its effect on regulating mitochondrial dynamics2. In agreement with previous findings of mitochondrial morphology in GSCs and NSTCs, we found that mitochondria fission was predominant in GSCs relative to NSTCs, as demonstrated by the increased percentage of fragmented mitochondria GSCs (Fig. 3a, b). In contrast, mitochondrial fusion was more frequently observed in NSTCs relative to GSCs, as demonstrated by increased percentage of tubular mitochondria in NSTCs (Fig. 3a, b). We then evaluated mitochondrial morphologies in CPT1A-overexpressed GSCs and the control GSCs, and found that CPT1A overexpression promoted mitochondrial fusion in GSCs (Fig. 3c, d). To address how CPT1A affected mitochondrial dynamics, we examined the expression of regulators governing mitochondrial dynamics. Immunoblotting results showed that CPT1A overexpression increased the phosphorylation of DRP1 at Ser-637 that has been implicated in maintaining mitochondrial fusion in GSCs (Fig. 3e). However, CPT1A overexpression had negligible impact on the expression of other mitochondria-regulating molecules including FIS1, MFF, MFN1, MFN2, or OPA1 (Fig. 3e). These data suggest that CPT1A overexpression in GSCs could promote mitochondrial fusion partially through promoting DRP1 phosphorylation at Ser-637.
a Immunofluorescent staining of TOM20 in GSCs and NSTCs of GBM1. b Mitochondrial morphology was evaluated from 150 cells of three different dishes in each group. c Immunofluorescent staining of TOM20 in CPT1A-overexpressed GSCs and control GSCs. d Mitochondrial morphology quantification of CPT1A-overexpressed GSCs and control GSCs. e Immunoblot analysis of CPT1A, DRP1, the activating phosphorylation of DRP1 at Ser-616, the suppressive phosphorylation of DRP1 at Ser-637, FIS1, MFF, MFN1, MFN2, and OPA1 in CPT1A-overexpressed GSCs and control GSCs. Experiments were independently performed for at least three times. White boxes show representative mitochondrial morphology and enlarged images were presented below. Scale bars, 4 μm. *p < 0.05 and **p < 0.01.
Silencing CPT1A in NSTCs promotes mitochondrial fission
We investigated the effect of CPT1A on regulating mitochondrial dynamics in NSTCs. Silencing CPT1A in NSTCs promoted mitochondrial fission, as demonstrated by the reduced percentage of tubular mitochondria in NSTCs expressing CPT1A-targeting shRNAs relative to the control NSTCs (Fig. 4a–d). Immunoblotting results confirmed that CPT1A silencing in NSTCs promoted the phosphorylation of DRP1 at Ser-616 that has been implicated to maintaining mitochondrial fission (Fig. 4e). Disruption of CPT1A activity by etomoxir, a small molecule inhibitor of CPT1A18, increased the percentage of fragmented mitochondria, confirming that disruption of CPT1A in NSTCs induced mitochondrial fission (Fig. 4f, g).
a Immunofluorescent staining of TOM20 in GBM1 NSTCs transduced by control or sh1-CPT1A and sh2-CPT1A lentiviral vector. b Mitochondrial morphology was evaluated from 150 cells of three different dishes in each group of GBM1. c Immunofluorescent staining of TOM20 in GBM2 NSTCs transduced by lentiviral control vector or sh1-CPT1A and sh2-CPT1A. d Mitochondrial morphology was evaluated from 150 cells of three different dishes in each group of GBM2. e Immunoblot analysis of CPT1A, DRP1, phospho-DRP1(Ser-616), and phopho-DRP1(Ser-636) in NSTCs derived from GBM1 and GBM2. Experiments were independently performed for at least three times. f Immunofluorescent staining of TOM20 in NSTCs derived from GBM1 treated with DMSO or ETO. g Mitochondrial morphology was evaluated from 150 cells of three different dishes in DMSO-treated NSTCs or ETO-treated NSTCs of GBM1. White boxes show representative mitochondrial morphology and enlarged images were presented below. Scale bars, 4 μm. *p < 0.05 and **p < 0.01.
Inhibition of DRP1 compromises the effect of CPT1A silencing on promoting mitochondrial fission and inducing GSC stemness
To investigate whether CPT1A regulated GBM cell stemness by DRP1-mediated mitochondrial dynamics, we used mdivi-1 as a DRP1-mediated mitochondrial fission inhibitor in NSTCs expressing CPT1A shRNAs or control NSTCs. CPT1A silencing significantly promoted mitochondrial fission of NSTCs, while mdivi-1 inhibited mitochondrial fission and promoted mitochondrial fusion in CPT1A-silencing NSTCs (Fig. 5a–d). Immunoblotting results confirmed that mdivi-1 treatment suppressed the phosphorylation of DRP1 at Ser-616 but up-regulated the phosphorylation of DRP1 at Ser-637 in CPT1A-silencing NSTCs (Fig. 5e), confirming that mdivi-1 reverses the effect of CPT1A disruption on mitochondrial dynamics. Furthermore, mdivi-1 treatment largely rescued the expressions of OLIG2, SOX2 and GFAP in CPT1A-silencing NSTCs (Fig. 5f). Together, these results indicated that inhibition of DRP1-mediated mitochondrial fission could rescue the effect of CPT1A silencing on promoting GBM cell stemness.
a Immunofluorescent staining of TOM20 in GBM1 NSTCs of expressing sh-CPT1A or control vector (NC) and those treated with DRP1 inhibitor mdivi-1. b Immunofluorescent staining of TOM20 in GBM2 NSTCs expressing sh-CPT1A or control vector and those treated with mdivi-1. c Mitochondrial morphology quantification of GBM1 NSTCs in each group. d Mitochondrial morphology quantification of GBM2 NSTCs in each group. e Immunoblot analysis of CPT1A, DRP1, phospho-DRP1 (Ser-616), and phopho-DRP1 (Ser-637) in NSTCs expressing sh-CPT1A or control vector and those treated with mdivi-1. f Immunoblot analysis of GFAP, OLIG2, and SOX2 in NSTCs. Experiments were independently performed for at least three times. White boxes show representative mitochondrial morphology and enlarged images were presented below. Scale bars, 4 μm. *p < 0.05 and **p < 0.01.
CPT1A overexpression inhibits GSC-derived xenograft growth and prolongs survival in tumor-bearing mice
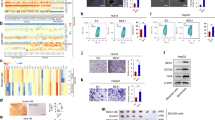
We then investigated whether enforced expression of CPT1A could suppress GSC-derived tumor growth. GSC-derived orthotopic xenografts were monitored using MRI scanning. We found that CPT1A overexpression in GSCs significantly inhibited GSC xenograft growth (Fig. 6a, b) and prolonged survival in tumor-bearing mice (Fig. 6c). H&E staining of tumor regions confirmed that CPT1A overexpression decreased xenograft size (Fig. 6d). Electron microscopy showed that xenograft cells overexpressing CPT1A exhibited more tubular mitochondria relative to control xenograft cells (Fig. 6e). Immunofluorescent staining demonstrated a reduced percentage of Ki67+ proliferating cells and SOX2+ GSCs in GSC xenografts overexpressing CPT1A (Figs. 6f, g). Together, these data demonstrate that CPT1A overexpression inhibits GSC-derived xenograft growth and prolongs animal survival.
a MRI-T2WI scanning of tumor-bearing mice on days 7, 13, and 19 after tumor implantation. b Quantification of xenograft tumor volume on day 19. Scale bars, 5 mm. c Kaplan–Meier survival analysis of mice bearing GBM1 GSC-derived xenografts. d Representative images of brain sections stained by hematoxylin and eosin (H&E) harvested on day 19 after tumor transplantation. Scale bars, 4 mm. e Electron microscopy images of mitochondria in tumor cells of GSC-derived xenograft transduced by lentiviral control vector or CPT1A. Mitochondrial morphology was evaluated from 600 cells of six tumors in each group of GBM1. Scale bars, 1 μm. f Immunofluorescent staining of Ki67 (red) in GSC xenografts. Scale bars, 25 μm. The percentage of Ki67+ cells was calculated from 60 randomized areas of six xenograft tumors in each group. g Immunofluorescent staining of SOX2 (red) in GSC xenografts. Scale bars, 25 μm. The percentage of SOX2+ cells was calculated from 60 randomized areas of six xenograft tumors in each group. *p < 0.05 and **p < 0.01.
Discussion
Our study demonstrates the important role of CPT1A in regulating mitochondrial dynamics and GSC differentiation. GSCs contribute to therapeutic resistance and tumor recurrence, hence eliminating GSC population has been proved with potential therapeutic benefit in preclinical studies4. Inducing GSC differentiation represents one of the promising GSC-eliminating strategies. We found that CPT1A inhibited the growth of GSC-derived xenograft, accompanied with promoting GSC differentiation and decreasing the population of GSCs, uncovering its therapeutic significance. Previous preclinical studies have identified several molecules for inducing GSC differentiation. Transcription factors FOXO3 and β-catenin can activate downstream AMP-activated protein kinase (AMPK) signaling to promote GSC differentiation19. Epigenetic regulations including miR-124, miR-137 and miR-145 have also been implicated to the regulation of GSC differentiation20. Furthermore, a histone deacetylase 3 inhibitor RGFP966 promoted GSC differentiation through inhibiting TGF-β signaling pathway21. Our research demonstrated that CPT1A is another promising target for inducing GSC differentiation.
Interestingly, we found CPT1A was highly expressed in NSTCs relative to GSCs both in human GBM specimens and in in vitro cultured cells. Several pathways might be involved in regulating the transcription of CPT1A during CSC differentiation. A previous work has showed that CREB-PGC1α pathway is pivotal for cell differentiation22. As CPT1A has been predicted as a downstream transcriptional target of CREB-PGC1α pathway23, it is theoretically possible that CREB-PGC1α pathway can induce CPT1A expression during cell differentiation. FGFR is also a crucial molecule for regulating CSC stemness and proliferation24. It has been reported that inhibition of FGFR promoted CSC differentiation, and blocking FGFR could induce CPT1A expression25, suggesting CPT1A is regulated by FGFR during CSC differentiation. The mechanism underlying the increased expression of CPT1A in NSTCs of GBM needs to be further clarified.
Although mitochondrial morphology differs between GSCs and NSTCs, the molecular link mediating mitochondria morphology and GSC stemness remains unclear. It has been reported that inhibition of mitochondrial fission promotes GSC differentiation13. Our data revealed that CPT1A regulated GSC mitochondrial dynamics by regulating DRP1 activity. DRP1 promotes mitochondrial fission through translocating on the outer membrane of mitochondria from cytosol and its enzymatic activity is mediated by the phosphorylation of Ser-616. The phosphorylation of Ser-637 in DRP1 reversed the process and promoted mitochondrial fusion9. It was reported that GSCs preferentially expressed cyclin-dependent kinase 526, which could activate DRP1 through phosphorylating Ser-616 and result in mitochondrial fission13. While in NSTCs, calcium/calmodulin-dependent protein kinase type 2 was preferentially expressed and inhibited DRP1 activity through phosphorylating Ser-637 of DRP1 to promote mitochondrial fusion13. We thus propose that the different activation state of DRP1 between GSCs and NSTCs are critical for regulating mitochondrial fusion and fission. Further investigations of the mechanisms underlying the GSC/NSTC states and mitochondrial dynamics (fusion and fission) are warranted. Several molecules may be involved in CPT1A regulation on mitochondrial dynamics. For example, AMPK could regulate CPT1A expression through AMPK/ACC/CPT1A pathway27, suggesting AMPK-CPT1A-DRP1 cascade may regulate mitochondrial dynamics in GSCs. In addition, it has been reported that CPT1A suppressed p53 phosphorylation to inhibit the downstream pathway activation28. As p53 has been demonstrated to regulate Drp1 phosphorylation29, p53 pathway might be one of the mediators of CPT1A-regulated DRP1 phosphorylation. Besides, CPT1A deficiency could inhibit ERK activation30, which is involved in the phosphorylation of Drp1 at Ser-61631. Therefore, MAPK/ERK pathway might also be involved in CPT1A-regulated DRP1 phosphorylation. Future works are warranted to investigate the pathways connecting CPT1A and DRP1 phosphorylation in GSCs.
Our results demonstrated that CPT1A overexpression inhibited GSC proliferation and the growth of GSC-derived xenografts, suggesting a GSC-suppressive role of CPT1A, which is consistent with that derived from colorectal cancers and breast cancers. Wiśniewski et al.32 identified CPT1A as a down-regulated protein in colorectal cancers by comparing the proteomes of normal colorectal enterocytes and colorectal cancer cells, which was in accordance with our observation. Overexpression of CPT1A could also inhibit the proliferation of breast cancer cells33. However, in nasopharyngeal carcinomas, up-regulation of CPT1A promoted radiation resistance23, indicating the tumor-promoting role of CPT1A. Furthermore, CPT1A overexpression increased mitochondrial ROS production and promoted cell proliferation of prostate cancers, in the presence of lipids and androgen deprivation conditions16. These studies suggest a tumor-context dependent role of CPT1A under different metabolic conditions. Further studies are needed to elucidate the underlying mechanism of CPT1A in regulating tumor progression of different tumors.
References
Janjua, T. I. et al. Frontiers in the treatment of glioblastoma: past, present and emerging. Adv. Drug Deliv. Rev. 171, 108–138 (2021).
McKinnon, C., Nandhabalan, M., Murray, S. A. & Plaha, P. Glioblastoma: clinical presentation, diagnosis, and management. BMJ 374, n1560 (2021).
Ma, Q. et al. Cancer stem cells and immunosuppressive microenvironment in glioma. Front. Immunol. 9, 2924 (2018).
Zhou, W. et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat. Cell Biol. 17, 170–182 (2015).
Peiris-Pages, M., Martinez-Outschoorn, U. E., Pestell, R. G., Sotgia, F. & Lisanti, M. P. Cancer stem cell metabolism. Breast Cancer Res. 18, 55 (2016).
Diehn, M. et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458, 780–783 (2009).
Shibao, S. et al. Metabolic heterogeneity and plasticity of glioma stem cells in a mouse glioblastoma model. Neuro. Oncol. 20, 343–354 (2018).
Chan, D. C. Mitochondrial dynamics and its involvement in disease. Annu. Rev. Pathol. 15, 235–259 (2020).
Fonseca, T. B., Sanchez-Guerrero, A., Milosevic, I. & Raimundo, N. Mitochondrial fission requires DRP1 but not dynamins. Nature 570, E34–E42 (2019).
Cassidy-Stone, A. et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell 14, 193–204 (2008).
Tabara, L. C., Morris, J. L. & Prudent, J. The complex dance of organelles during mitochondrial division. Trends Cell Biol. 31, 241–253 (2021).
Lee, W. T. & St, J. J. The control of mitochondrial DNA replication during development and tumorigenesis. Ann. NY Acad. Sci. 1350, 95–106 (2015).
Xie, Q. et al. Mitochondrial control by DRP1 in brain tumor initiating cells. Nat. Neurosci. 18, 501–510 (2015).
Schlaepfer, I. R. & Joshi, M. CPT1A-mediated fat oxidation, mechanisms, and therapeutic potential. Endocrinology 161, bqz046 (2020).
Tan, Z. et al. Targeting CPT1A-mediated fatty acid oxidation sensitizes nasopharyngeal carcinoma to radiation therapy. Theranostics 8, 2329–2347 (2018).
Joshi, M. et al. CPT1A Over-expression increases reactive oxygen species in the mitochondria and promotes antioxidant defenses in prostate cancer. Cancers (Basel) 12, 3431 (2020).
Shi, Y. et al. Tumour-associated macrophages secrete pleiotrophin to promote PTPRZ1 signalling in glioblastoma stem cells for tumour growth. Nat. Commun. 8, 15080 (2017).
O’Connor, R. S. et al. The CPT1a inhibitor, etomoxir induces severe oxidative stress at commonly used concentrations. Sci. Rep. 8, 6289 (2018).
Song, J. E. et al. Mitochondrial fission governed by drp1 regulates exogenous fatty acid usage and storage in hela cells. Metabolites 11, 322 (2021).
Silber, J. et al. MiR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 6, 14 (2008).
Liu, M. Y. et al. Molecular hydrogen suppresses glioblastoma growth via inducing the glioma stem-like cell differentiation. Stem Cell Res. Ther. 10, 145 (2019).
Xing, F. et al. The Anti-Warburg effect elicited by the cAMP-PGC1alpha pathway drives differentiation of glioblastoma cells into astrocytes. Cell Rep. 18, 468–481 (2017).
Du, Q. et al. PGC1alpha/CEBPB/CPT1A axis promotes radiation resistance of nasopharyngeal carcinoma through activating fatty acid oxidation. Cancer Sci. 110, 2050–2062 (2019).
Quan, M. Y. et al. An FGFR/AKT/SOX2 signaling axis controls pancreatic cancer stemness. Front. Cell Dev. Biol. 8, 287 (2020).
Song, H. et al. Metabolic flexibility maintains proliferation and migration of FGFR signaling-deficient lymphatic endothelial cells. J. Biol. Chem. 297, 101149 (2021).
Mukherjee, S. et al. CDK5 inhibition resolves PKA/cAMP-independent activation of CREB1 signaling in glioma stem cells. Cell Rep. 23, 1651–1664 (2018).
Yao, Q. et al. Yin Zhi Huang, a traditional Chinese herbal formula, ameliorates diet-induced obesity and hepatic steatosis by activating the AMPK/SREBP-1 and the AMPK/ACC/CPT1A pathways. Ann. Transl. Med. 8, 231 (2020).
Teng, H. et al. Fatty acid degradation plays an essential role in proliferation of mouse female primordial germ cells via the p53-dependent cell cycle regulation. Cell Cycle. 15, 425–431 (2016).
Yuan, Y. et al. P53/Drp1-dependent mitochondrial fission mediates aldosterone-induced podocyte injury and mitochondrial dysfunction. Am. J. Physiol. Renal. Physiol. 314, F798–F808 (2018).
Han, S. et al. CPT1A/2-mediated FAO enhancement- a metabolic target in radioresistant breast cancer. Front. Oncol. 9, 1201 (2019).
Kashatus, J. A. et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell. 57, 537–551 (2015).
Wisniewski, J. R. et al. Absolute proteome analysis of colorectal mucosa, adenoma, and cancer reveals drastic changes in fatty acid metabolism and plasma membrane transporters. J. Proteome Res. 14, 4005–4018 (2015).
Aiderus, A., Black, M. A. & Dunbier, A. K. Fatty acid oxidation is associated with proliferation and prognosis in breast and other cancers. BMC Cancer. 18, 805 (2018).
Funding
This research was supported by grants from the National Key Research and Development Program of China (2017YFC0110105) and National Natural Science Foundation of China (81821003, 81922056).
Author information
Authors and Affiliations
Contributions
Conception and design: X.-W.B., Y.-F.P., and Y.S. Cell culturing: M.L., H.Z., and Y.-Q.L. Staining: M.L., Y.Q.L., C.-H.L., and Z.-C.H. Immune blotting: M.L., H.Z., Q.L., and W.-Y.W. Animal experiments: Y.-Q.L., X.-N.Z., C.C., and M.M. PCR: X.-Q.C., W.-J.F., and Q.N. Analysis of data: K.-D.Y. and C.W. Writing of the manuscript: X.-W.B., Y.-F.P., and Y.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The human GBM section staining was approved by the Ethics Committees of Southwest Hospital (KY2021124). The animal use was approved by the Ethics Committees of Southwest Hospital (AMUWEC20211956).
Consent for publication
All authors agreed on the manuscript and publication.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Luo, M., Liu, YQ., Zhang, H. et al. Overexpression of carnitine palmitoyltransferase 1A promotes mitochondrial fusion and differentiation of glioblastoma stem cells. Lab Invest 102, 722–730 (2022). https://doi.org/10.1038/s41374-021-00724-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00724-0
This article is cited by
-
Drp1: Focus on Diseases Triggered by the Mitochondrial Pathway
Cell Biochemistry and Biophysics (2024)
-
Carnitine palmitoyltransferase 1A promotes mitochondrial fission by enhancing MFF succinylation in ovarian cancer
Communications Biology (2023)