Abstract
Osteocytes are mechanosensitive cells that control bone remodeling in response to mechanical loading. To date, specific signaling pathways modulated by mechanical loading in osteocytes are not well understood. Yes associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ), the main effectors of the Hippo pathway, are reported to play a role in mechanotransduction and during osteoblastogenesis. Here, we hypothesized that YAP/TAZ signaling mediates osteocyte mechanosensing to target genes of the bone remodeling process. We aimed to investigate the contribution of YAP/TAZ in modulating the gene expression in an osteocyte-like cell line MLO-Y4. We developed a 3D osteocyte compression culture model from an MLO-Y4 osteocyte cell line embedded in concentrated collagen hydrogel. 3D-mechanical loading led to the increased expression of mechanosensitive genes and a subset of chemokines, including M-csf, Cxcl1, Cxcl2, Cxcl3, Cxcl9, and Cxcl10. The transcription regulators YAP and TAZ translocated to the nucleus and upregulated their target genes and proteins. RNAseq analysis revealed that YAP/TAZ knockdown mediated the regulation of several genes including osteocyte dendrite formation. Use of YAP/TAZ knockdown partially blunted the increase in M-csf and Cxcl3 levels in response to MLO-Y4 compression. These findings demonstrate that YAP/TAZ signaling is required for osteocyte-like cell mechano-transduction, regulates the gene expression profiles and controls chemokine expression.
Similar content being viewed by others
Background
Mechanical loading plays an important role in maintaining bone homeostasis by the regulation of bone remodeling1. Osteocytes, the most abundant cell type embedded in the mineralized bone matrix, represent the main mechanosensors that control bone remodeling in response to mechanical forces2,3. Osteocyte dendrites within the canaliculi networks allow sensing of mechanical loading and communicate with other bone cell types to regulate their activities2. Osteocyte mechanosensing properties result in the activation of different types of receptors and different intracellular signaling pathways3. Indeed, loading induces osteocyte responses associated with early uptake of intracellular calcium via ion channels, hemi-channels (mainly Connexin-43) and primary cilia4. Moreover, loading promotes calcium influx in osteocytes, together with the production of adenosine triphosphate, nitric oxide and prostaglandin E2, the latter being able to activate bone formation and inhibit bone resorption5,6,7,8,9,10,11,12,13,14. In addition to these metabolites, osteocytes produce factors that regulate the sequence of events comprising the process of bone remodeling. Hence, osteocytes are the main source of receptor activator of nuclear factor kappa-Β ligand (RANK-L) and can negatively regulate osteoclast differentiation in response to mechanical loading by downregulating the ratio of RANK-L to osteoprotegerin12,13,15. Osteocyte mechanosensing also downregulates the production of sclerostin that subsequently activates bone formation following mechanical forces16,17,18. Among the signaling pathways implicated in mechanotransduction, mechanical loading rapidly activates Wnt/β-catenin signaling in osteocytes via a prostaglandin-mediated mechanism19.
Yes associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ) were first discovered in Drosophila and described as the main regulators of organ growth during development20,21. YAP/TAZ is regulated by mechanical and cytoskeletal cues and acts as a mechanotransducer controlling cell fate in response to the properties of the cellular microenvironment22,23. Indeed, large adhesive area, stiff matrix, low cell density or mechanical pressure led to the nuclear translocation of YAP/TAZ, thus enabling the control of cell proliferation, differentiation and apoptosis24. Nuclear YAP/TAZ acts as a transcriptional co-activator because it can interact with transcription factors, mainly the members of the TEA domain transcription factor family (TEAD)25. YAP/TAZ activity is controlled at multiple levels, notably via its phosphorylation, which induces its cytoplasmic or membrane shuttling, and subsequent degradation by the proteasome26. These regulations are mediated directly by cellular contact, matrix properties or soluble factors and indirectly by YAP/TAZ phosphorylation mediated by the core Hippo kinase cassette, mammalian Ste20-like ½ (MST1/2) and large tumor suppressor kinase ½ (LATS1/2).
YAP/TAZ interacts with different signaling pathways that are implicated in osteoblastogenesis, such as the transforming growth factor β/ bone morphogenetic protein pathway and Wnt/β-catenin signaling27,28,29. Emerging evidence suggests that YAP/TAZ plays a role in mechanosensitivity during osteoblastogenesis. YAP/TAZ has been shown to induce the commitment of mesenchymal stem cells toward the osteoblast lineage by inhibiting adipogenesis and chondrogenesis, particularly in response to fluid shear stress or low cell density30,31,32,33. Microgravity induces a decrease in osteoblastogenesis owing to the downregulation of TAZ activity34. Li and collaborators also demonstrated in the osteocytic cell line MLO-Y4 that Piezo mediates mechanotransduction and induces YAP/TAZ nuclear translocation in response to shear stress35. Finally, YAP/TAZ seemingly has a role in osteocytes in vivo in that the deletion of YAP/TAZ using dentin matrix acidic phosphoprotein 1 (DMP1-Cre) in mice induced low bone mass characterized by increased osteoclast activity and decreased bone formation36.
In light of these findings, the question of a potential role of YAP/TAZ in osteocytes has been raised, particularly in response to 3D mechanical loading. Therefore, the aim of this study was to characterize the response of the osteocyte-like cell line MLO-Y4 following mechanical loading using a new 3D culture model subjected to compressive stress.
Materials and methods
Cell lines cultures and mechanical stress experiments
MLO-Y4 cells were cultured on collagen-coated flasks previously rinsed to reach pH 7.0 before plating cells (rat tail tendon type I collagen, 0.15 mg/mL in 0.02 M acetic acid solution) in alpha minimum essential medium (αMEM, GIBCO) supplemented with 5% heat-inactivated fetal bovine serum (GIBCO), 5% heat-inactivated calf serum (Hyclone), 100 U/mL penicillin and 100 mg/mL streptomycin and grown at 37 °C in 5% CO2 as described37.
2D cultures and mechanical stretch
For the preparation of the 2D MLO-Y4 cultures subjected to mechanical stretch, MLO-Y4 cells were treated with trypsin and plated at 1.2 × 105 cells/well in BioFlex Culture Plates coated with collagen type I (Flexcell International Corp., Dunn Labortechnick GmbH, Asbach, Germany). BioFlex Culture Plates are 6-well plates containing a flexible silicone rubber membrane at the bottom of each well. MLO-Y4 cells were grown at 37 °C in a humidified incubator containing 5% CO2. At 80% confluence, 24 hr before stretching, the medium was replaced with fresh medium containing a reduced amount of serum (1% heat-inactivated fetal bovine serum and 1% heat-inactivated calf serum). MLO-Y4 cells grown on BioFlex plates were submitted to an equibiaxial mechanical stretch using the Flexcell FX-5000T Tension System (Flexcell International Corp., Burlington, NC, USA), a computer-regulated bioreactor that apply cyclic or static stretch to 2D cells culture models by applied regulated vacuum pressure below each silicone flexible rubber membrane38,39. Percentage of stretch, frequency, form of signal and duration of stretch are adjustable. The 2D MLO-Y4 cultures were submitted to an equi-biaxial dynamic stretch with a sinusoidal waveform at 0.3 Hz and 3% elongation for 9 hr at 37 °C and 5% CO2. Frequency and elongation settings used were previously described40. Control cultures plates were prepared and underwent the same conditions and were kept in un-stretched conditions. Gene expression analyses were performed at 0 hr post-stretch. 2D experiments were repeated at least 4-times.
3D cultures and mechanical compression
MLO-Y4 cells were embedded in concentrated collagen type I hydrogel (rat tail tendon type I collagen 4 mg/mL in 0.5 M acetic acid solution, 0.02 mM NaOH, PBS and αMEM 1X) at 2 × 106 cells/mL. Cells were cultured for 14 days in complete medium in a humidified atmosphere at 37 °C and 5% CO2 to allow the formation of an osteocyte-like structure with dendrites elongation across collagen hydrogel before loading experiments. Collagen gels with diameter 13-mm underwent compression according to the usage specifications of the BioPress Compression Plates with the Flexcell FX-5000C Compression System (Flexcell International Corp., Dunn Labortechnick GmbH, Asbach, Germany). The BioPress Compression Plates consist of 6-well plates containing a flexible silicone rubber membrane at the bottom. On each membrane a rigid plastic piston is fixed and a circular foam disc attached to each plastic piston confines the samples to the central region of each well. Each collagen gel is put on the plastic piston, into the circular foam disc. In total, 3 ml culture medium is introduced into each well and a stationary platen closes each well. The 3D MLO-Y4 culture transferred in BioPress was compressed by using the Flexcell FX-5000C Compression System, a computer-regulated bioreactor that regulates positive air pressure to compress tissue samples or 3D cell cultures in vitro41. Briefly, a force applied by positive air pressure below each flexible rubber membrane causing the deformation of membrane leads to compression of the 3D MLO-Y4 culture between the piston and the stationary platen. This system allows the pressure to be uniform in the sample area. At 24 hr before compression, the medium was replaced with fresh medium containing reduced amount of serum (1% heat-inactivated fetal bovine serum and 1% heat-inactivated calf serum), and 3D MLO-Y4 cultures were submitted to cyclic compression of 0–40 kPa with a sinusoidal waveform at 1 Hz for 9 hr in a humidified atmosphere at 37 °C and 5% CO2. This load condition (40 kPa and 1 Hz) had already been tested on type I collagen gel and enabled a cellular response to mechanical stress, so we used this condition for our model42. The frequency of 1 Hz corresponds to the frequency of a moderate walk43. Control 3D MLO-Y4 cultures were kept in unloaded conditions and were not placed into the device. At the end of the experiment, collagen hydrogels were fixed with AntigenFix for 24 hr or hydrolyzed at 37 °C with 5 mg/mL type I collagenase solution (Sigma) for RNA and protein extraction. 3D experiments were repeated at least three times.
All mechanical stress experiments outlined in this study were conducted using systems located in the Mechanobiology core facility of BioMedTech Facilities at INSERM US36 - CNRS UMS2009 - Paris Descartes.
Isolation of total RNA, reverse transcription, and real-time quantitative PCR
Isolation of total RNA involved using RNA Isolate II Minikit (Bioline) according to the instructions of the manufacturer. The mRNA isolated from each sample was reverse transcribed by using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystem) from 1 μg total RNA. Real-time quantitative PCR involved using cDNA (1:40 final dilution per reaction) with Sensifast (Bioline). Tata binding protein (TBP) was used for normalization, and mRNA expression was calculated as percentage of TBP expression (2−∆CT (mean gene - mean gene reference)) to give an indication of the level of expression. The primer sequences are in Supplementary Table 1.
Production and transfection of shRNA
Lentiviral shRNA particles were produced from the 293 T cell lines by using the Ready-to-Use Lentiviral Packaging (CPCP-K2A Cellecta) and shRNA plasmid from Santa Cruz Biotechnology (TAZ shRNA plasmid sc-38569-SH, YAP shRNA plasmid sc-38638-SH and shControl# sc-108060). Transfection of 293 T involved using 2 million cells (2.65 × 104 cell/cm2) with 10 µg lentiviral packaging, 30 µL of Lipofectamine 2000 (Thermofisher) and 2 µg shRNA plasmid. Lentiviral particles were collected 24 hr after transfection and applied on MLO-Y4 cells previously plated at 1.6 × 104 cells/cm2. MLO-Y4 were infected for 48 hr and selected with puromycine at 10 µg/mL for at least one week.
RNA-sequencing (RNAseq) analysis
RNAseq analysis involved using duplicates for the following conditions: shControl# loaded and unloaded, shYAP loaded and unloaded, shTAZ loaded and unloaded. The preparation of strand-specific cDNA libraries and the sequencing on an Illumina HiSeq4000 sequencer in 150 bp paired-end mode was performed by Eurofins GATC Biotech GmbH, Konstanz, Germany.
Quality controls (QCs)
A quality check of the FASTQ files containing the raw reads was involved using the software package FastQC v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The software returns a comprehensive HTML report containing several modules that summarize key details of interest, such as GC content, duplication level and adapter content. For each module, there is a fail/warn/pass icon that allows the detection of any potential problems before the downstream analysis, thus avoiding any false positive results from the beginning. All of the 24 FASTQ files provided successfully passed nearly every module of the quality test and therefore were considered suitable for the next steps of the pipeline. The FastQC reports were then aggregated into 3 MultiQC v1.8 (https://multiqc.info/) reports for ease of visualization and summary purposes44. The MultiQC reports were generated by using Python 3.7.4.
Pseudoalignment
Kallisto v0.46.1 represents an innovative tool for transcript abundance quantification45. In Kallisto, the raw sequence reads (i.e., FASTQ files) are directly compared to transcript sequences and then used to quantify transcript abundance. The comparison of the sequencing reads to the transcripts involves using a transcriptome de Bruijn graph46 in which each node is a k-mer (i.e., a unique string of k nucleotides from a set of sequences). Therefore, each node is associated with a transcript or set of transcripts formally described as the k-compatibility class of that vertex. The alignment is based on the intersection of k-compatibility classes while skipping the redundant k-mers along a path (i.e., equivalence classes), thus speeding up the process. A number of 100 bootstraps were used for all samples. The output was represented by an abundance file together with a summary of the pseudo-alignment. The programming language used was Python 3.7.4 and the mapping was done on the mouse transcriptome mm10.
Differential gene expression (DGE) analysis
The tximport Bioconductor package (release 3.10) was used to import and summarize the transcript-level abundance estimates from the previous pseudo alignment step. Tximport estimates un-normalized counts and transcript lengths and summarizes them into matrices for use with downstream gene-level analysis packages47. To determine whether any genes were enriched in different experimental setups, the estimated count matrices obtained after the import were studied by using DESeq2 v1.26.0, an R-based tool that performs DGE analysis by using a negative binomial distribution48. DESeq2 generates a DESeqDataSet (dds) to store read counts and the intermediate estimated quantities during statistical analysis. The differential analysis steps are automatically performed by calling the DESeq function, and the results tables are generated by calling the results function, the log2 fold change depending on the factor levels specified. To visualize gene changes, an unsupervised gene clustering analysis was applied with the CRAN heatmap package (v.0.12). The level of clustering was also investigated by a principal component analysis (PCA) plot after regularizing the original count data with the regularized log function (rlog)49. Lists of highly up/downregulated genes were created, with absolute log2FoldChange ≥ log2(1.5) and P-adjusted value (padj) ≤0.05 as cut-off criteria. MA (ratio intensity) plots were also generated. R v3.6.1 was used for the analysis.
Gene ontology (GO) analysis
Functional annotations of the lists of significantly enriched genes corresponding to the experimental conditions were studied by using goseq, a Bioconductor package that takes length bias into account by calculating a Probability Weighting Function (PWF)50,51. Goseq-based analysis requires a vector containing all the genes found in an RNAseq dataset for an experimental comparison, the genes being separated in a binary manner: enriched ones (1), uninteresting ones (0). A Wallenius distribution is then used to determine whether each GO category is over/under-represented among the set of differentially expressed genes. The output GO terms were sorted based on their associated p values (p ≤ 0.05 as cut-off), and their biological meaning was studied by using the online AMIGO2 tool (http://amigo.geneontology.org/amigo/landing). The annotations are based on the Ensembl genome browser.
Western blot analysis
Proteins were separated by using 4–12% Novex Tris-Glycine mini gel electrophoresis (Fisher Scientific) under reducing conditions and transferred onto a PVDF membrane. Blotted membranes were blocked by using 5% skin milk at room temperature for 1 hr, incubated overnight at 4 °C with primary antibody diluted in 5% bovine serum albumin and incubated with horseradish peroxidase-conjugated secondary antibody diluted in 5% skin milk for 1 hr at room temperature. Chemiluminescent detection was achieved with Clarity Western ECL Substrat (BioRad). Primary antibodies were mouse anti-YAP (1:1000, sc-101199, Santa Cruz Biotechnology), mouse anti-TAZ (1:1000, 560235, BD Bioscience), rabbit anti-YAP/TAZ (1:1000, 8418 S, Cell signaling) and anti-GAPDH as a control (1:1000, ab9484, Abcam). Secondary antibodies were rabbit anti-mouse (1:10000, ab6728, Abcam) and goat anti-rabbit (1:10000, ab6721, Abcam). Chemiluminescent signals were detected by using the Azure Imaging System and were quantified with ImageJ.
Histology
Collagen hydrogels were fixed with AntigenFix for 24 hr and embedded in paraffin; 5-µm thick sections were obtained by using an automatic microtome (Leica) For histological cryosections, left femurs from 6-month-old mice were fixed in 4% paraformaldehyde overnight at 4 °C and then decalcified in 0.5 M EDTA (pH 7.4) at 4 °C with constant shaking for 10 days with four changes of EDTA solution. Decalcified bones were washed twice with a mixture of 10% sucrose in phosphate buffered saline (PBS) and then with 30% sucrose in PBS overnight at 4 °C. The next morning, all samples were cryoprotected with a 50/50 mixture of 30% sucrose solution and then 100% OCT compound overnight at 4 °C. Next day, bones were embedded with 100% OCT compound at −20 °C and cryosections of 5-μm thick were obtained by using a cryostat (X, Thermo Scientific).
For immunofluorescence analysis from collagen hydrogel, 5-µm thick sections were dewaxed and treated with HIER Citrate Buffer pH 6.0 (Zymoted) for 1 hr at 95 °C, followed by treatment with 0.1% pepsin diluted in 10% acetic acid solution for 1 hr at 37 °C and with glycin 1% for 30 min at room temperature. Permeabilization involved using 0.1% PBS-Triton for 10 min at room temperature and blocking epitopes was performed in PBS-1% BSA-1% glycine. Primary antibodies used for immunofluorescence are listed in the previous section. For immunofluorescence analysis from femur, 5-µm thick sections were permeabilized with Tween 0.05% and then blocked with PBS-serum donkey serum 5%-glycine 100 mM for 30 min.
All antibodies used were diluted in PBS-BSA-1%-glycine-1% for collagen hydrogel and PBS-serum donkey serum 5%-glycine 100 mM for femurs with a dilution ratio of 1:200 for primary antibodies (rabbit anti-YAP/TAZ, 8418 S, Cell Signaling or purified rabbit IgG, Rb-003-V, ImmunoReagents) and 1:100 for secondary antibodies coupled to fluorochrome DyLight550 (donkey anti-rabbit, SA5-10039, Invitrogen). DAPI staining was used to identify nuclei. The fluorescent signal was preserved by using DAKO mounting fluorescence medium. Images were acquired by using the Apotome optical sectioning system (Zeiss) with an inverted microscope (Zeiss Axio Observer Z1).
Statistical analysis
Data are presented as mean ± SEM and dot plots represent each collagen hydrogel. Statistical analysis involved using the Mann-Whitney U test for comparing two independent samples. Two-way ANOVA was used for multiple comparisons and controlling the false discovery rate correction. P < 0.05 was considered statistically significant. Statistical analyses involved using GraphPad Prism 7.00 (GraphPad Software, La Jolla, CA, USA).
Results
3D mechanical loading enhances the mechanosensing of the osteocyte-like cell line MLO-Y4
To define the best conditions for osteocyte-like cell line culture in response to mechanical loading, we used both cyclic mechanical 2D stretching and 3D compression on MLO-Y4 cells. In comparison to 2D stretching and unloaded mechanical conditions, 3D compression activated certain mechanosensitive genes such as E11 and COX2-encoding Ptgs2 (Fig. 1A). Therefore, the 3D osteocyte compression model was chosen for all the following experiments.
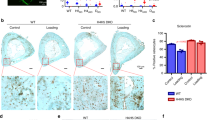
A E11/gp38 and Ptsg2 (encoding COX-2) gene expression assessed by real-time qPCR in response to mechanical loading in 2D (stretching) or 3D culture (compression) (B) YAP/TAZ target genes expression assessed by real-time qPCR after mechanical loading (C) YAP/TAZ nuclear translocation measured from Apotome immunofluorescence images of YAP/TAZ (in green) and nuclei (DAPI, in blue). Scale bars: 20 µm. Graphic represent the percentage of nuclear YAP/TAZ positive cells. Each dot plot represents individual collagen hydrogel obtained from three different experiments. D YAP1 and WWTR1 (TAZ) gene expression assessed by real-time qPCR in response to mechanical loading (E) Immunoblotting of YAP and TAZ from MLO-Y4 submitted to mechanical loading in 3D culture. TAZ immunoblotting was performed once. YAP quantification involved three different experiments. Data are mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001 by Mann–Whitney test or two-way ANOVA.
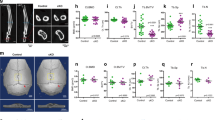
Osteocyte-like cell line MLO-Y4 mechanical loading led to activation of the YAP/TAZ pathway, as shown by an increase in the expression of the target genes Ankdr1 and Tead4 (Fig. 1B). The activation of YAP/TAZ signaling was further confirmed by immunofluorescence with quantification of the nuclear translocation, this being increased in response to mechanical loading (Fig. 1C). 3D compression not only reduced Yap expression but also increased Taz expression (Fig. 1D). Moreover, 3D compression enhanced the protein expression of YAP/TAZ, thus suggesting a stabilization of the YAP/TAZ protein (Fig. 1E). To confirm YAP/TAZ expression in vivo in osteocytes, antibody labeling was performed in femurs of 6-month-old mice. YAP/TAZ staining was found in both cortical and trabecular bones (Fig. 2).
The images YAP/TAZ immunostaining in trabecular and cortical bone were obtained from 6-month-old wild-type mice. YAP/TAZ (in green) and nuclei (in blue) staining was detected with the Apotome optical sectioning system (Zeiss, France). White arrow represents YAP/TAZ-positive osteocytes. Scale bars: 100 µm.
Characterization of YAP/TAZ gene response to mechanical loading in the MLO-Y4 osteocyte-like cell line
RNAseq was used to determine the biological processes modulated by YAP/TAZ as part of the osteocyte mechanosensing process. The screening was performed on cells with shRNA knockdown of Yap/Taz. Yap and Taz shRNA-based silencing was sufficient to decrease both YAP/TAZ mRNA and protein levels (Table 1, Supplementary Fig. 1A, B).
RNAseq analysis demonstrated via principal component analysis that regardless of the YAP/TAZ knockdown, the loading component accounted for most of the variance (principal component 1 = 92%), which suggests a strong effect of the loading factor (Fig. 3A). Figure 3B–D shows the MA plots that are the plots of log averages (A values) against log2 fold changes (M values). These were generated as part of the RNAseq analysis, and the most up/downregulated genes specific to each experimental condition are indicated. Cxcl2 was the most highly upregulated gene induced by mechanical loading in control cells (Fig. 3B). The 3D loading induced a higher number of enriched genes (Fig. 3B) as compared with YAP/TAZ deletion, thus eluding to a discrete gene profile triggered by the silencing of these transcriptional coactivators. The gene response of the deletion of YAP or TAZ under mechanical loading led to some similarity, thus suggesting a certain level of redundancy. Fbn2 was one of the common genes upregulated by both YAP and TAZ individual deletion in osteocytic cells under mechanical loading. In addition, the silencing of YAP and TAZ was effective because both transcriptional cofactors were among the most downregulated genes (Fig. 3C, D).
A Principal component analysis (PCA) for mechanical loading (principal component 1 [PC1]) and YAP/TAZ silencing (principal component 2 [PC2]). Each dot represents a sample and each color the conditions. B–D Scatter plots describe the distribution of changes between two conditions (on the y axis: (B) shControl unloaded vs shControl loaded; (C) shControl loaded vs shYAP loaded; (D) shControl loaded vs shTAZ loaded) compared to the average normalized read counts (on the x axis). Colored dots represent genes that are significantly expressed (padj ≤ 0.05). Down- and upregulated genes are indicated in red and green, respectively.
Gene ontology (GO) analysis
GO analysis was used to identify biological processes regulated by YAP/TAZ under mechanical loading in the osteocyte-like cell line MLO-Y4. The output GO terms resulting from the goseq analysis pipeline were sorted based on their p value, and details are provided for the top most-enriched terms of every experimental comparison (Supplementary Tables 2, 3). Supplementary Table 2 indicates that TEAD1 and TEAD2 were among the top genes regulated in response to mechanical loading via YAP because YAP deletion highlighted an alteration of the genes contributing to these GO terms. Several GO terms related to dendritic morphogenesis were highlighted after cell loading, thus suggesting the contribution of YAP and TAZ in the relation between osteocytic cell functioning and the lacuna-canaliculi system (Supplementary Tables 2, 3).
YAP/TAZ mediates mechanicall -induced chemokine expression in the MLO-Y4 osteocyte-like cell line
Because Cxcl2 was the most upregulated gene in MLO-Y4 osteocytes after mechanical loading, a part of the RNAseq analysis focused on the effect of mechanical loading on chemokine expression (Supplementary Table 3). Mechanical strains induced a large increase in Cxcl1, Cxcl2, Cxcl3, Cxcl9, Cxcl10, and Csf1 levels in control cells (shRNA control). However, the deletion of YAP and TAZ abolished the increase in Cxcl3, Cxcl9, Cxcl10, and Csf1 levels (Supplementary Table 3). To confirm the RNAseq results related to chemokine expression in terms of YAP/TAZ modulation, we used real-time qPCR analyses. Mechanical loading indeed increased level of Csf1 (M-CSF), Cxcl3, Cxcl9, and Cxcl10 in control cells (Fig. 4). Silencing YAP/TAZ partially blunted the upregulation of Csf1 and Cxcl3 after mechanical loading. In contrast, Cxcl10 and Cxcl9 upregulation in response to mechanical loading did not depend on YAP/TAZ.
Discussion
In this study, we developed an innovative 3D osteocyte-like cell-line culture compression system with MLO-Y4 cells to explore the gene modifications induced by mechanical loading and to identify whether YAP/TAZ signaling acts as a mechanosensitive mediator in the cells. Our experimental model allowed us to study osteocyte mechanotransduction in a 3D culture consisting of collagen type I, thus representing a more physiologically relevant environment. This model also allowed for applying mechanical compression levels that recapitulate the different forces acting upon the bone, such as tension, fluid shear and compressive forces52. As a result, the osteocytic cell response following compression in our experimental condition was amplified as compared with 2D cultures.
Here we provide the first piece of evidence of activated YAP/TAZ signaling in response to 3D mechanical loading in the MLO-Y4 osteocyte cell line. Our results and those of Li and collaborators demonstrate that YAP/TAZ signaling was activated by both 3D compression and fluid shear stress in the MLO-Y4 cell line35.
Indeed, YAP/TAZ activation in response to shear stress was previously described in mesenchymal stem cells or osteoblastic cells and recently MLO-Y4 cells32,35,53. In vivo YAP/TAZ deletion using DMP1-Cre mice decreased bone mass, associated with increased osteoclast activity and decreased osteoblastogenesis, thus suggesting an major contribution of YAP/TAZ in mature osteoblasts and osteocytes36. This phenotype could be due to the failure of osteoblasts to fully differentiate into mature osteocytes, thus leading to a blockage of osteoblasts at a differentiation state associated with a high potential to induce osteoclastogenesis. To date, there was no specific model for YAP/TAZ deletion pertaining exclusively to osteocytes and aiming to investigate the role of YAP/TAZ in osteocyte mechanotransduction. Here, we found that YAP/TAZ mediates the mechanically induced expression of chemokines such as M-CSF and Cxcl3 in osteocyte-like cells. We further confirmed YAP/TAZ expression in osteocytes in vivo in femurs of 6-month-old mice. Additional studies are required to confirm our in vitro results, notably YAP/TAZ and chemokine expression in osteocytes in mice submitted to hypergravity or microgravity. Here, we used the MLO-Y4 cell line that is reported to not express sclerostin, a regulator of the Wnt/β-catenin pathway important for osteocytes, but no other cell line showed the whole expression profile of primary osteocytes. This is a limitation of our study because we could not exclude that sclerostin regulates and impact the mechanically induced chemokine expression. Also, the Wnt/β-catenin pathway is closely related to YAP/TAZ signaling and we could not rule out that sclerostin, normally upregulated by mechanical loading, limits YAP/TAZ activation in osteocytes.
Our GO analysis suggested that YAP and TAZ could be implicated in osteocyte canaliculi formation. This finding is consistent with recent literature references demonstrating that YAP and TAZ deletion, by means of 8kb-DMP1-Cre mice, impaired osteocyte perilacunar/canaliculi remodeling54. Also, YAP/TAZ deletion in osteoblasts impaired bone mechanical properties and matrix collagen content55, but we did not find an effect of YAP/TAZ on collagen or matrix protein expression. These discrepancies could be due to the shortcomings of our in vitro model that does not exactly mimic the in vivo bone matrix environment.
Emerging data show that osteocytes produce a large panel of chemokines in response to fluid shear stress or cyclic compressive forces, as demonstrated in the MLO-Y4 cell line by microarray or RNAseq56,57,58. Here, mechanical loading upregulated the mRNA expression of Cxcl1, Cxcl2, Cxcl3, Cxcl9, and Cxcl10. Others showed that Cxcl1, Cxcl2, and Cxcl9 were able to induce migration, proliferation and adhesion of bone marrow-derived macrophages59,60,61,62. A recent study demonstrated that Cxcl10 secreted by osteocytes induced osteoclastogenesis, and a disruption of this regulation may be involved in osteoporosis63. In addition, M-CSF, a well-known cytokine known to induce osteoclast precursor proliferation, was upregulated in our experiments involving mechanical loading64. Our results suggest that osteocytes could represent a source of chemokines released in response to mechanical loading to trigger the homeostatic activity of bone effector cells, because osteocytes were demonstrated to produce chemokines such as Cxcl10, which, in turn, induce osteoclastogenesis63. The results also raise the question of the dialogue between osteocyte and muscle under loading because osteocytes were found to secrete chemokines that modulate myogenesis65. Further studies are needed to fully describe the role of the osteocyte secretome modulated by YAP/TAZ in response to loading and its ability to modulate the activity of bone, muscle or other tissues.
In conclusion, our results provide the first piece of evidence that YAP/TAZ is activated in osteocyte-like cells after 3D mechanical stimulation and regulates chemokine expression. Our results suggest that YAP/TAZ represents a modulator of mechanotransduction in osteocyte-like cells. We also demonstrate that osteocytic MLO-Y4 cells are a major source of different chemokines, notably Cxcl3, Cxcl9, and Cxcl10, released in response to mechanical loading.
Data availability
Datasets will be available from the authors upon reasonable request.
References
Robling, A. G., Castillo, A. B. & Turner, C. H. Biomechanical and molecular regulation of bone remodeling. Annu. Rev. Biomed. Eng. 8, 455–498 (2006).
Schaffler, M. B., Cheung, W.-Y., Majeska, R. & Kennedy, O. Osteocytes: master orchestrators of bone. Calcif. Tissue Int. 94, 5–24 (2014).
Xiao, Z. & Quarles, L. D. Physiological mechanisms and therapeutic potential of bone mechanosensing. Rev. Endocr. Metab. Disord. 16, 115–129 (2015).
Yavropoulou, M. P. & Yovos, J. G. The molecular basis of bone mechanotransduction. J. Musculoskelet Neuronal. Interact 16, 221–236 (2016).
Taylor, A. F. et al. Mechanically stimulated osteocytes regulate osteoblastic activity via gap junctions. Am. J. Physiol. Cell Physiol. 292, C545–C552 (2007).
Cherian, P. P. et al. Mechanical strain opens connexin 43 hemichannels in osteocytes: a novel mechanism for the release of prostaglandin. Mol. Biol. Cell 16, 3100–3106 (2005).
Genetos, D. C., Kephart, C. J., Zhang, Y., Yellowley, C. E. & Donahue, H. J. Oscillating fluid flow activation of GAP junction hemichannels induces ATP release from MLO-Y4 osteocytes. J. Cell Physiol. 212, 207–214 (2007).
Bao, L., Locovei, S. & Dahl, G. Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett. 572, 65–68 (2004).
Fox, S. W., Chambers, T. J. & Chow, J. W. Nitric oxide is an early mediator of the increase in bone formation by mechanical stimulation. Am. J. Physiol. 270, E955–E960 (1996).
Zaman, G. et al. Mechanical strain stimulates nitric oxide production by rapid activation of endothelial nitric oxide synthase in Osteocytes. J. Bone Miner. Res. 14, 1123–1131 (1999).
Tan, S. D. et al. Osteocytes subjected to fluid flow inhibit osteoclast formation and bone resorption. Bone 41, 745–751 (2007).
Liao, C. et al. Shear stress inhibits IL-17A-mediated induction of osteoclastogenesis via osteocyte pathways. Bone 101, 10–20 (2017).
You, L. et al. Osteocytes as mechanosensors in the inhibition of bone resorption due to mechanical loading. Bone 42, 172–179 (2008).
Kulkarni, R. N., Bakker, A. D., Everts, V. & Klein-Nulend, J. Inhibition of osteoclastogenesis by mechanically loaded osteocytes: Involvement of MEPE. Calcif. Tissue Int. 87, 461–468 (2010).
Xiong, J., Piemontese, M., Onal, M. & Campbell, J. Osteocytes, not osteoblasts or lining cells, are the main source of the RANKL required for osteoclast formation in remodeling bone. PLoS ONE 10, e0138189 (2015).
Robling, A. G. et al. Mechanical stimulation of bone in vivo reduces osteocyte expression of Sost/sclerostin. J. Biol. Chem. 283, 5866–5875 (2008).
Tu, X. et al. Sost downregulation and local Wnt signaling are required for the osteogenic response to mechanical loading. Bone 50, 209–217 (2012).
Spatz, J. M. et al. The Wnt-inhibitor sclerostin is up-regulated by mechanical unloading in Osteocytes in-vitro. J. Biol. Chem. 290, 16744–16758 (2015).
Lara-castillo, N. et al. In vivo mechanical loading rapidly activates β -catenin signaling in osteocytes through a prostaglandin mediated mechanism. Bone 76, 58–66 (2015).
Dong, J. et al. Elucidation of a universal size-control mechanism in drosophila and mammals. Cell 130, 1120–1133 (2007).
Pan, D. Hippo signaling in organ size control. Genes Dev. 21, 886–897 (2007).
Halder, G., Dupont, S. & Piccolo, S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 13, 591–600 (2012).
Hansen, C. G., Moroishi, T. & Guan, K.-L. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 25, 499–513 (2015).
Panciera, T., Azzolin, L., Cordenonsi, M. & Piccolo, S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Publ. Gr. 18, 758–770 (2017).
Kim, M., Jang, J.-W. & Bae, S.-C. DNA binding partners of YAP/TAZ. BMB Rep. 51, 126–133 (2018).
Piccolo, S., Dupont, S. & Cordenonsi, M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev. 94, 1287–1312 (2014).
Morgan, J. T., Murphy, C. J. & Russell, P. What do mechanotransduction, Hippo, Wnt, and TGF β have in common? YAP and TAZ as key orchestrating molecules in ocular health and disease. Exp. Eye Res. 115, 1–12 (2013).
Azzolin, L. et al. YAP/TAZ incorporation in the B-catenin destruction complex orchestrates the Wnt response. Cell 158, 157–170 (2014).
Varelas, X. et al. The hippo pathway regulates Wnt/B-catenin signaling. Dev. Cell 18, 579–591 (2010).
Hong, J. et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Sci. Rep. 309, 1074–1078 (2005).
Tang, Y. & Weiss, S. J. Snail/Slug-YAP/TAZ complexes cooperatively regulate mesenchymal stem cell function and bone formation. Cell Cycle 16, 399–405 (2017).
Kim, K. M. et al. Shear stress induced by an interstitial level of slow flow increases the osteogenic differentiation of mesenchymal stem cells through TAZ activation. PLoS ONE 9, e9427 (2014).
McBeath, R., Pirone, D. M., Nelson, C. M., Bhadriraju, K. & Chen, C. S. Cell shape, cytoskeletal tension, and RhoA regulate stemm cell lineage commitment. Dev. Cell 6, 483–495 (2004).
Chen, Z., Luo, Q., Lin, C. & Song, G. Simulated microgravity inhibits osteogenic differentiation of mesenchymal stem cells through down regulating the transcriptional co-activator TAZ. Biochem. Biophys. Res. Commun. 468, 21–26 (2015).
Li, X. et al. Stimulation of Piezo1 by mechanical signals promotes bone anabolism. Elife 8, e49631 (2019).
Xiong, J., Almeida, M. & O’Brien, C. A. The YAP/TAZ transcriptional co-activators have opposing effects at different stages of osteoblast differentiation. Bone 112, 1–9 (2018).
Kato, Y., Windle, J., Koop, B., Mundy, G. & Bonewald, L. Establishment of an Osteocyte-like Cell Line, MLO-Y4. Am. Soc. Bone Miner. Res 12, 2014–2023 (1997).
Banes, A. J., Gilbert, J., Taylor, D. & Monbureau, O. A new vacuum-operated stress-providing instrument that applies static or variable duration cyclic tension or compression to cells in vitro. J. Cell Sci. 75, 35–42 (1985).
Gilbert, J. A., Weinhold, P. S., Banes, A. J., Link, G. W. & Jones, G. L. Strain profiles for circular cell culture plates containing flexible surfaces employed to mechanically deform cells in vitro. J. Biomech. 27, 1169–1177 (1994).
Hens, J. R. et al. TOPGAL mice show that the canonical Wnt signaling pathway is active during bone development and growth and is activated by mechanical loading in vitro. J. Bone Min. Res. 20, 1103–1113 (2005).
Fermor, B. et al. The effects of static and intermittent compression on nitric oxide production in articular cartilage explants. J. Orthop. Res. 19, 729–737 (2001).
Hara, M. et al. Construction of collagen gel scaffolds for mechanical stress analysis. Biosci. Biotechnol. Biochem. 78, 458–461 (2014).
Sardroodian, M., Madeleine, P., Voigt, M. & Hansen, E. A. Freely chosen stride frequencies during walking and running are not correlated with freely chosen pedalling frequency and are insensitive to strength training. Gait Posture 42, 60–64 (2015).
Ewels, P., Magnusson, M., Lundin, S. & Max, K. Data and text mining MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic rna-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Compeau, P. E. C., Pevzner, P. A. & Tesler, G. Why are de Bruijn graphs useful for genome assembly? Nat. Biotechnol. 29, 987–991 (2017).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Lever, J., Krzywinski, M. & Altman, N. Principal component analysis. Nat. Methods 14, 641–642 (2017).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 (2010).
Oshlack, A. & Wakefield, M. J. Transcript length bias in RNA-seq data confounds systems biology. Biol. Direct. 4, 14 (2009).
Stoltz, J. et al. Influence of mechanical forces on bone: introduction to mechanobiology and mechanical adaptation concept. J. Cell. Immunother. 4, 10–12 (2018).
Kaneko, K., Ito, M., Naoe, Y., Lacy-Hulbert, A. & Ikeda, K. Integrin αv in the mechanical response of osteoblast lineage cells. Biochem. Biophys. Res. Commun. 447, 352–357 (2014).
Kegelman, C. D. et al. YAP and TAZ mediate osteocyte perilacunar/canalicular remodeling. J. Bone Min. Res. 35, 196–210 (2020).
Kegelman, C. D. et al. Skeletal cell YAP and TAZ combinatorially promote bone development. FASEB J. 32, 2706–2721 (2018).
Govey, P. M. et al. Integrative transcriptomic and proteomic analysis of osteocytic cells exposed to fluid flow reveals novel mechano-sensitive signaling pathways. J. Biomech. 47, 1838–1845 (2014).
Govey, P. M., Imamura, Y. & Donahue, H. J. Mapping the osteocytic cell response to fluid flow using RNA-Seq. J. Biomech. 48, 4327–4332 (2015).
Chen, W. et al. Gene expression patterns of osteocyte-like MLO-Y4 cells in response to cyclic compressive force stimulation. Cell Biol. Int. 34, 425–432 (2010).
Ha, J. et al. CXC chemokine ligand 2 induced by receptor activator of NF-kB ligand enhances osteoclastogenesis. J. Immunol. 184, 4717–4724 (2010).
Ha, J., Lee, Y. & Kim, H. H. CXCL2 mediates lipopolysaccharide-induced osteoclastogenesis in RANKL-primed precursors. Cytokine 55, 48–55 (2011).
Hardaway, A. L., Herroon, M. K., Rajagurubandara, E. & Podgorski, I. Marrow adipocyte-derived CXCL1 and CXCL2 contribute to osteolysis in metastatic prostate cancer. Clin. Exp. Metastasis 32, 353–368 (2015).
Kwak, H. B. et al. Monokine induced by interferon-gamma is induced by receptor activator of nuclear factor kappa B ligand and is involved in osteoclast adhesion and migration. Blood 105, 2963–2969 (2005).
Liu, P. et al. Loss of menin in osteoblast lineage affects osteocyte-osteoclast crosstalk causing osteoporosis. Cell Death Differ. 24, 672–682 (2017).
Amarasekara, D. S. et al. Regulation of osteoclast differentiation by cytokine networks. Immune Netw. 18, 1–18 (2018).
Wood, C. L., Pajevic, P. D. & Gooi, J. H. Osteocyte secreted factors inhibit skeletal muscle differentiation. Bone Rep. 6, 74–80 (2017).
Acknowledgements
We thank Dr Lynda F Bonewald for providing the MLO-Y4 osteocyte cell line. We thank Dr Kristina Kampmann from GATC Biotech for RNAseq experiments and analysis. We also thank Laura Smales for excellent editing of the paper.
Funding
The study was supported by SYBIL European consortium.
Author information
Authors and Affiliations
Contributions
Conception and design (Z.M., E.F., S.J.M., R.F., H.E., C.S.M.); analysis and interpretation of the data (Z.M., S.D., H.E., C.S.M.); drafting of the article (Z.M., S.D., E.F., S.J.M., H.E., C.S.M.); critical revision of the article for important intellectual content (Z.M., S.D., S.J.M., H.E., C.S.M.); final approval of the article (all authors); provision of study materials or patients (E.F., R.F., H.C., C.S.M.); statistical expertize (Z.M., D.S.); obtaining funding (C.S.M.); administrative, technical, or logistical support (E.F., H.C., B.M., R.F.); collection and assembly of data (Z.M., C.S.M.).
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zarka, M., Etienne, F., Bourmaud, M. et al. Mechanical loading activates the YAP/TAZ pathway and chemokine expression in the MLO-Y4 osteocyte-like cell line. Lab Invest 101, 1597–1604 (2021). https://doi.org/10.1038/s41374-021-00668-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00668-5
This article is cited by
-
Genetic interactions between polycystin-1 and Wwtr1 in osteoblasts define a novel mechanosensing mechanism regulating bone formation in mice
Bone Research (2023)
-
The Multifaceted Effects of Osteocytic TGFβ Signaling on the Skeletal and Extraskeletal Functions of Bone
Current Osteoporosis Reports (2023)