Abstract
The placenta is the first organ to form and performs the functions of the lung, gut, kidney, and endocrine systems. Abnormalities in the placenta cause or reflect most abnormalities in gestation and can have life-long consequences for the mother and infant. Placental villi undergo a complex but reproducible sequence of maturation across the third-trimester. Abnormalities of villous maturation are a feature of gestational diabetes and preeclampsia, among others, but there is significant interobserver variability in their diagnosis. Machine learning has emerged as a powerful tool for research in pathology. To capture the volume of data and manage heterogeneity within the placenta, we developed GestaltNet, which emulates human attention to high-yield areas and aggregation across regions. We used this network to estimate the gestational age (GA) of scanned placental slides and compared it to a baseline model lacking the attention and aggregation functions. In the test set, GestaltNet showed a higher r2 (0.9444 vs. 0.9220) than the baseline model. The mean absolute error (MAE) between the estimated and actual GA was also better in the GestaltNet (1.0847 weeks vs. 1.4505 weeks). On whole-slide images, we found the attention sub-network discriminates areas of terminal villi from other placental structures. Using this behavior, we estimated GA for 36 whole slides not previously seen by the model. In this task, similar to that faced by human pathologists, the model showed an r2 of 0.8859 with an MAE of 1.3671 weeks. We show that villous maturation is machine-recognizable. Machine-estimated GA could be useful when GA is unknown or to study abnormalities of villous maturation, including those in gestational diabetes or preeclampsia. GestaltNet points toward a future of genuinely whole-slide digital pathology by incorporating human-like behaviors of attention and aggregation.
Similar content being viewed by others
Introduction
The placenta is the first organ to form and functions as the fetal lung, gut, kidney, endocrine, and immune systems. As an active participant in gestation, it consumes as much oxygen at term as the entire fetus [1]. Placental pathology causes and reflects adverse events in pregnancy [2, 3]. Pathology in the placenta can have lifelong consequences for mothers and offspring, including increased risk of cardiovascular disease [4], bronchopulmonary dysplasia [5], cerebral palsy [6], colorectal carcinoma [7], and asthma [8]. Therefore, the examination of the placenta can yield considerable benefit. Yet, <20% of placentas are examined in the United States, and significant lesions are frequently unrecognized [9, 10].
Digital pathology has the potential to revolutionize our understanding of placental function and disease [11]. Routine diagnostic pathology relies on qualitative assessment and pattern recognition. Research studies on human placentas usually rely on these assessments or quantitative measurements of selected regions done by hand. A more quantitative, thorough examination may identify new biology and pathophysiology. The sheer volume of archived glass slides of placentas, ~120,000 at our institution alone, with ~500,000 cells in each whole-slide image (WSI), provides an enormous untapped reservoir of material for hypothesis development and testing.
In comparison, clinical examination captures only a fraction of the information from each slide, and the quality is dependent on the examiner. Despite the accessibility of placentas at the time of birth, that information is discarded in most cases. Once an AI system is operating, increasing the scale, adding new populations or diseases is simple. This could include placentas from low-resource or international settings, patients with specific sociodemographic factors, or patients with emerging diseases of pregnancy, like COVID-19.
Changes over time
Over the course of the second- and third-trimesters, the placental disc increases approximately tenfold in size. The most significant microscopic changes are within the terminal villi, with increased numbers of small villi with decreased cellularity, increased stromal density, migration of capillaries to below the syncytial membrane, and collection of syncytiotrophoblast nuclei into knots. These changes have the overall effect of minimizing the distance between maternal and fetal blood [12,13,14]. In analogy with the lung, this results in maximum surface area with minimum diffusion distance for oxygen and nutrients (Fig. 1). Determination of the appropriateness of villous maturation is a key step in assessing a placenta. This task is daunting, as it involves the integration of the factors mentioned above across multiple slides to form a single gestalt. Accordingly, interobserver variability is high [15,16,17].
In the early 3rd-trimester (1, 3), syncytiotrophoblast (ST) nuclei are evenly spaced. Capillaries (C) are distant from maternal blood, which bathes the villi. The stroma consists of loose extracellular matrix proteins with frequent macrophages and fibroblasts (brown and pink stars). At term (2, 4), the villi are smaller. Syncytiotrophoblast nuclei are gathered into knots (K), thinning the vasculosyncytial membrane. Capillaries are directly beneath the syncytiotrophoblast layer. Stroma is denser with lower cellularity.
Gestational age (GA) is the single most important factor in perinatal well-being. The probability of a newborn successfully transitioning from womb to nursery to home increases markedly with GA, and the probability of adverse outcomes including hypoxic-ischemic encephalitis, necrotizing enterocolitis, and bronchopulmonary dysplasia markedly decrease [18]. Accurate identification of GA most commonly relies on sonographic measurements made in the first- or second-trimester [19,20,21]. These measurements may not be available in low-resource settings or when prenatal care is inadequate. Other methods, such as the recalled date of the last menstrual period or sonographic measurements made in the third-trimester, are less accurate.
The placenta and digital pathology
Compared to neoplasia, the placenta is relatively understudied by digital pathology. Studies using photomicrographs of single fields and manual annotation show the potential for scientific discovery using deep, image-based phenotyping of the placenta. Manual measurement of villous and vascular surface area has shown changes over pregnancy [12, 13]. Preeclampsia (PreE) has been associated with changes in villous count, area, diameter, capillary count, and degree of capillarization in the villous core [14]. Gestational diabetes has been associated with decreased villous vascular volume [22]. Abnormal villous maturation has a genetic expression signature—placentas with a diagnosis of accelerated maturation have gene expression more appropriate for placentas delivered 4.7 weeks later with normal maturation [23].
More recent studies support the feasibility of applying modern machine learning and digital pathology techniques to the placenta. Studies have shown the ability to segment villi from scanned slides and measure their stromal density and vessel numbers [24, 25]. Published algorithms exist for identifying cytotrophoblast, fibroblast, macrophage, syncytiotrophoblast, and vascular endothelial cells in the placenta [26].
Deep learning models employing convolutional neural networks (CNN) have shown impressive performance for identifying image content in multiple domains and tasks, including digital pathology [27,28,29,30,31,32]. In training, networks commonly learn to associate a single image or HPF to an outcome or finding of interest. Contrary to CNN’s implicit assumption of one image corresponding to one label, a single WSI contains thousands of HPF with considerable heterogeneity. Practicing pathologists must examine all HPF, attend to fields they consider representative, and aggregate their findings to produce a single diagnosis. The gap between algorithm development and practice reduces the clinical relevance of many AI studies including those in the broader medical imaging field. We propose an algorithm that learns the patient outcome from a collection or set of images in training. This helps to incorporate more regions from each WSI during the learning procedure.
The problem of aggregation extends beyond digital pathology and is present whenever a model receives multiple inputs. Practitioners must decide at which stage of the pipeline data are incorporated, how they are weighted, and the extent to which aggregation is trainable. In non-image tasks, data are routinely input as a single vector allowing complex trainable interactions. Conversely, ensemble strategies may aggregate results from multiple separately trained models without back propagation. Choices in aggregation strategy are liable to be suboptimal if practitioners are unaware that a choice is being made.
This study aims to develop a deep learning model that incorporates and predicts across whole slides and demonstrates the utility of that model in the estimation of GA in placenta—a low concordance task in notoriously heterogeneous tissue.
Materials, subjects, and methods
Patients and materials
Pathology reports from patients delivering 1/1/2010 to 10/31/2019 were retrieved from the laboratory information system (Cerner Build List Id: 2014.08.1.36). GA, clinical history, and diagnoses, including accelerated, delayed, and appropriate maturation, were extracted using regular expressions (6.2) and the Natural Language Toolkit (NLTK, version 3.3) on Python (version 3.6.9) as described [33, 34].
We identified cases with an obstetrically determined GA of 24–42 weeks with an original pathologic diagnosis of appropriate villous maturation, confirmed through a review by a practicing perinatal pathologist (JAG). This GA was considered the ground truth for each case.
Clinical examination of placentas at our institution includes 1 cassette of membranes, 1 of umbilical cord sections, 1 with three incisional biopsies of the placental disc’s maternal surface (basal plate plus villi), 2 cassettes of the representative non-lesional full-thickness placental disc, and additional cassettes containing any lesions. The maternal surface biopsies and full-thickness sections are selected from the inner 2/3 of the radius of the placental disc were reviewed for possible scanning. We selected a slide with morphology consistent with clinically determined GA without mass-forming lesions or villous abnormalities. Given low counts in the earliest GA, we allowed cases with decidual or chorionic plate pathology (e.g., chorioamnionitis).
One slide per patient with villous tissue, either basal villous wedges or full-thickness placental disc, was selected and scanned at the institutional Pathology Core Facility using a Hamamatsu Nanozoomer 2.0 HT scanner at ×20 objective magnification. 154 slides were split randomly, stratified by GA, into training, validation, and test sets with proportions of ~70% (107 slides), ~15% (23 slides), and ~15% (24 slides), respectively. Because deliveries are not evenly distributed across the GA and maturation anomalies are more prevalent at earlier GA, the training, validation, and test sets are not precisely balanced at each GA. A list of cases and corresponding GA is presented in Supplementary Table 1.
Regions of terminal villi with villous maturation consistent with GA were box annotated by the pathologist. Stem villi, areas of fibrin deposition, and septae were avoided. On full-thickness sections, parabasal areas were preferentially annotated. In total, 1918 region annotations (at least 10 per slide) were made. Regions were extracted with OpenSlide (1.1.1) on Python (3.6.9) and were color normalized using the method from Macenko et al. [35]. Regions were tiled into 512 × 512 pixel high-power fields (HPF) at ×20 magnification level and shrunk to 256 × 256 (effective magnification ×10), for a total of 26,555 HPF (Supplementary Table 1). During training, HPF are augmented by random rotations and changes in brightness and contrast [36].
Baseline model
HPF are input into a feature extraction CNN based on VGG19 (30) with trainable weights initialized by a pre-trained model on ImageNet [37] in Keras (Tensorflow 2.3.0). The network is modified by replacing the fully connected layers in the original VGG19 architecture with a single fully connected layer of size 1024 with ReLU activation function and a dropout with a rate of 0.5. The extracted feature map is submitted to the representation learning sub-network, which consists of sequential fully connected layers of size 1024 and 256 with ReLU activation functions and a dropout with a rate of 0.5 after the first fully connected layer, and one linear node at the end to produce a single value—the estimated gestational age (EGA). The mean squared error loss between EGA and clinically determined GA (as the ground truth) is used to train the model. The baseline model was trained for 2000 epochs. To aggregate across a WSI for inference, the median EGA for all HPF is determined post hoc.
GestAltNet–input–glimpsing
In the base model, training explicitly links the clinical outcome to a single HPF. We propose an alternative network for estimating GA, GestAltNet (Figs. 2 and 3). GestAltNet learns in aggregate from a collection of images and relates the clinical outcome to a set of HPF during training. While the baseline model trains using a single HPF as input, GestAltNet uses a glimpse as input in training. Each glimpse consists of 16 randomly selected HPF from a single WSI, generally representing multiple regions. Glimpses are examined in batches of 64 and consumption of all batches represents one epoch. HPF and glimpses are resampled as needed to maintain glimpse and batch sizes. HPF are randomly assigned to glimpses at initialization and after every 50 epochs (chosen based on the performance in the validation set).
ROIs are tiled into HPFs (2nd panel, black lines). HPFs are randomly sampled without replacement across all ROIs of each patient to form a glimpse (third panel, HPF shading indicates glimpse) second panel from left, colored HPFs indicate their corresponding glimpse. Glimpses are constant size (16) except the last glimpse (purple oval) which takes the remainder. Glimpses from one patient are distributed across batches (fourth panel, gray ovals are glimpses from other patients).
Intermediate outputs (red boxes) are input to an attention sub-network. Features maps (f1–fn) are weighted by their attention (a1–an) and aggregated via weighted averaging (oval). The representation learning subnetwork estimates the gestational age (GA) based on the aggregated feature map f. The mean squared error (GA - GA)2 inside a total batch of 64 glimpses is used in backpropagation. The whole learning procedure is done in an end-to-end manner.
GestAltNet–pipeline–attention and aggregation
As in the baseline model, images are input into a VGG19 derived network. The intermediate output of VGG19 at block3, consisting of 256 3 × 3 kernels (Fig. 3, red squares), is input to the attention sub-network. This sub-network is a feedforward neural network with two fully connected layers of size 256, 256 with ReLU activation functions, a dropout with a rate of 0.5 after the first fully connected layer [38], and one linear node at the end. The linear node results in a single scalar value for each HPF in the glimpse, representing its attention. To limit extreme values, attentions are transformed using softmax.
A single aggregate feature map (f in Fig. 3) is obtained through weighted averaging over the feature maps of the 16 HPF within the glimpse, where weights are the corresponding HPF attentions. The aggregate feature map is submitted to the representation learning sub-network as in the baseline network to compute EGA. During training, mean squared error between EGA and clinically determined GA (ground truth) is used as the loss function, and backpropagation is performed end-to-end across the entire network. GestAltNet was trained for 500 epochs. For the whole-slide inference, the median EGA, computed across glimpses, was determined.
Metrics
To assess the overall accuracy, we measured the coefficient of determination (r2) and the absolute error in weeks. For test and unannotated slides, EGA was calibrated using the linear regression of EGA vs. GA for validation regions and whole slides (respectively). We considered an absolute error of >3 weeks as clinically significant because (1) accelerated villous maturation has been diagnosed based on an apparent GA of ≥37 weeks with chronologic GA of ≤34 weeks, i.e., 3 weeks [39]; (2) gene expression study showing accelerated villous maturation equates to 4.7 weeks ahead, and delayed maturation equates to 1.5 weeks behind normal gestation (average 3.1 weeks) [23]; (3) Using the placental weight reference of Pinar et al. [40], a placenta of average weight at one GA is considered large or small for gestational age (LGA, SGA) 3–5 weeks earlier or later. For example, a placenta with the mean weight for 24 weeks, 189 grams, is considered LGA at 21 weeks (expected 114–172 grams) and SGA at 27 weeks (expected 192–305 grams).
Attention and whole-slide estimation of GA
For the whole-slide level inference 36 new slides, neither previously annotated nor part of the training, validation and testing sets were used. The non-tissue area of the WSI was masked out by first applying Gaussian smoothing to the slide’s grayscale thumbnail, and then applying Otsu’s image binarization method to the thumbnail [41]. Attention was determined and GA was estimated on a per-HPF basis for all HPF. To determine appropriate attention thresholds for the selection of representative HPF in WSI level inference, we examined the per-HPF attention and accuracy over the non-overlapping HPF inside the tissue area of the WSI in our validation set. We set the lower threshold at the median attention of HPF with absolute errors of ≤3 weeks and the upper threshold at the 99th percentile of attention for HPF with absolute errors of ≤3 weeks in the validation set.
For generating heat maps, 87.5% overlapping HPF were extracted, and attention and EGA values were produced on a per-HPF basis. Attention was colored with minimum and maximum values scaled based on variation in the validation set. EGA was colored as H&E (appearing pink at low power) for absolute error ≤3 weeks, red if >3 weeks high and blue if >3 weeks low.
This study was approved by the institutional review board (STU00211333). WSI are available upon execution of a data use agreement.
Results
Interobserver variability
29,943 placentas were examined over 9.5 years by eight pathologists. Given a GA determined by clinical parameters, pathologists diagnose whether maturation is appropriate, accelerated, or delayed for the stated GA. Overall, 17,806 (60%) placentas were diagnosed with appropriate maturation, 5108 (17%) with accelerated maturation and 1024 (3.4%) with delayed maturation (Fig. 4). 6005 placentas (20%) received multiple diagnoses, for example, “appropriate for GA with regionally delayed maturation,” or had no description of maturation, which may occur when maturation is obscured by other findings like chorangiosis or post-mortem changes. The percentage of cases diagnosed as normal varied from 51 to 77%, as accelerated from 8.2 to 27%, and as delayed from 0.2 to 13%. Assuming a random distribution of placentas among pathologists, this represents significant interobserver variability.
Deep learning model performance
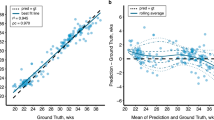
In the test set, the GestaltNet and baseline models showed r2 of 0.9444 and 0.9220, respectively (Fig. 4a–b). After calibration, the mean absolute error (MAE) was 1.0847 weeks for the GestaltNet model and 1.4505 for the baseline model. An error of ≥3 weeks is significant in evaluating GA. By this standard, both the GestaltNet and baseline models adequately estimated GA 24/24 test cases (Fig. 5).
Attention and estimation of GA across whole slides
The GestaltNet technique simulates a pathologist’s cognitive process of incorporating information across multiple regions of interest. However, it still relies on hand-annotated regions of interest selected to include representative, high-quality areas of tissue. To explore variation across tissue and emulate the pathologist attention and gestalt formation process across the whole slide, we obtained attention and EGA across 36 WSI that were unannotated and not part of the existing training, validation, or test sets. This resulted in an r2 of 0.8859 and an MAE of 1.3671 weeks. The model estimated GA was within 3 weeks of the actual GA in 35/36 (97.22%) cases (Fig. 6). To illustrate and further examine how WSI attention and prediction relate, we generated whole-slide attention and predictions for one WSI using overlapping HPF (Fig. 7). Perhaps surprisingly, given that we did not train our model to discriminate between different regions of the placenta, terminal villi show the highest attention, while stem villi, basal plate, and chorionic plate showed lower attention. GA estimation was variable within the villous region; however, the most accurate areas tended to be away from large stem villi or other masses. Some non-villous areas, including chorionic vessels, are attended to with divergent and inaccurate predictions.
Terminal villi are primarily high attention (yellow, regions 1, 5, and 6). Basal plate (left side of WSI and region 2), stem villi (region 3, intermixed with villous areas) and chorionic plate (right side of WSI and region 4) are generally low attention (purple). Estimated gestational age shows variegation with accurate areas (region 1) intermixed with areas with inaccurate low (blue, region 2) and high (red, region 3) estimates. Areas with low attention are disregarded (grayscale). The model is not explicitly trained to recognize tissue types and shows erroneous high attention to some areas. For example, one chorionic plate vessel (region 4) is part high- and part low-attention. The attended part of the vessel wall gives an estimate that misses low. Intravascular blood is attended and misses high.
Discussion
GA is the most significant factor in neonatal well-being. However, practicing pathologists rely on GA derived from other factors and show considerable inter-rater variability even in identifying whether the villous appearance is appropriate for the stated GA. We show that GA can be predicted with extraordinary accuracy from the beginning of viability (24 weeks) to post-term (42 weeks) using a deep learning approach. In practice, pathologists examine several regions across multiple whole slides, looking for different features that are either concordant or discordant with the chronological GA.
Developing a model for this task requires a solution to what we call “The Problem of Aggregation.” Our solution is to analyze multiple HPF in a glimpse. Aggregation occurs at the feature map stage. Feature maps are weighted based on the attention generated by an independent multilayer perceptron. The model takes the form of a single end-to-end network in which all sub-networks are trainable. We show that the integration of image features at an early stage with weighting and end-to-end trainability provides superior accuracy compared to post hoc averaging used in the baseline model. The improvement is highlighted by the stress test of calculating EGA without the regularization provided by human annotation.
One of the characteristics of deep learning algorithms that has made them so successful in digital pathology is their end-to-end learning approach. These adaptive algorithms learn to predict labels directly from pixel values in contrast to prior approaches that seek to incorporate a-priori knowledge in algorithm design. The unbiased end-to-end learning method is often credited as enabling deep learning models to learn latent predictive features in histology that may not be appreciated by human pathologists, but at the cost of algorithm interpretability.
End-to-end learning becomes practically difficult when labels correspond to an entire slide or a large region rather than a high-power field due to the scale of data corresponding to a single label and the limitations of computer hardware used to train deep learning algorithms. In this scenario, end-to-end learning requires that the mechanism for aggregating over multiple fields be incorporated into the learning model and be adaptive. In applications like tumor detection, a single positive field gives the whole-slide label, and have been solved using approaches like multiple instance learning. Other applications may be more compositional, requiring the interpretation and weighting of several tissue patterns, or learning to perform a weighted averaging over regions of the slide.
This paper provides a solution involving exhaustive random sampling of HPF representing a single case with the differential weighting of HPF by attention. This strategy is broadly applicable to any scenario when large amounts of data are consumed for each sample. However, it is particularly relevant for image analysis, where the interpretation of one portion of the image depends on context from other portions. For example, a pedestrian waving to another pedestrian on the other side of a street is more likely to enter the street than one waving to a departing car. In pathology, injured liver adjacent to a liver tumor represents mass effect, not cirrhosis. GestAltNet assigns attention weights on a per-HPF basis. This reflects the variability in information content between HPF, even within human-annotated ROI. Within-image attention, for example Grad-CAM, has been proposed to address the problem of interpretability in AI [42]. Theoretically, our attention could be used in a similar fashion, analogous to the use of dotting pens in pathology practice to annotate key areas for diagnosis. Within-image attention has been criticized for focusing on edges or complex structures and using similar patterns of attention to explain correct and incorrect answers [43]. It is not clear that a by-HPF system, such as GestAltNet, is immune from this problem, and the observation that it assigns similar attention to correct, miss-high, and miss-low regions (Fig. 7) is concerning.
Our choice of a single end-to-end network is also appealing in that it reflects human cognition, and all operations are potentially trainable. This mimics human thought patterns of aggregating impressions rather than diagnoses. Features may also be a more worthy area of focus as they are representations of biological phenomena, while HPF is arbitrary grids imposed by computer memory limitations.
Other authors have addressed the aggregation problem in the placenta with success. Clymer et al. use the multiple-resolution pyramid of images found in scanned slide files to identify vessels within placental membranes followed by clustering to produce a slide-level diagnosis as either containing healthy or pathologic maternal vessels [44]. However, this study did not use end-to-end training.
The future
This is among the first studies using machine learning in placental pathology and demonstrates the potential of this field. The extremely high accuracy in detecting normal morphology across gestation will allow the classification of many abnormalities, some currently unknown or with too low interobserver reliability to be useful.
In high-resource settings, GA is usually determined by first-trimester ultrasound. The system demonstrated is unlikely to replace this method but could be useful in cases where the dating of the pregnancy is unclear, or there is a discrepancy between the stated and apparent GA. In low or middle-income settings, photomicrographs of relevant areas taken using a smartphone and adapter could be used in lieu of WSIs [45]. In this use-case of human-machine cooperation, the small size of captured images means that a cloud-based network could provide estimated GA in real-time.
Accelerated and delayed villous maturation are among the most commonly reported placental findings in large data sets [33]. Nonetheless, they show poor inter-rater reliability, decreasing the significance of these findings. AI could be used in a quality assurance/improvement paradigm to improve interobserver variability in practice and is likely useful in identifying maturation abnormalities.
Our solutions to the problem of aggregation, as used in GestAltNet, will have applications far beyond the placenta. Intratumoral heterogeneity complicates neoplasia classification and is a marker for adverse outcomes [46,47,48]. In other non-neoplastic diseases, such as idiopathic pulmonary fibrosis, heterogeneity itself may be a criterion [49]. Beyond digital pathology, attention and aggregation within large and complex images remain fundamental challenges of image analysis.
Limitations
From a generalizability standpoint, the most significant limitations of this work are the use of a single site with consistent protocols and a single pathologist reviewer. Further work is necessary to develop and demonstrate generalizability across institutions and practitioners. Our demonstration of interobserver variability is limited in that pathologists are not reviewing the same placenta, but rather placentas submitted more or less randomly from the same population. The remainder of this work suggests that human-machine collaboration to overcome this variability will be more productive than perseverating on the precise degree of heterogeneity.
Conclusion
In conclusion, we report the machine learning-based estimation of GA from scanned histologic slides of the placenta. This demonstrates the tractability of this system and may be useful in diagnostic, quality, and research settings. We present a novel aggregation and attention model to manage and utilize the vast quantity of data present in whole slides.
Data availability
Data are available after the execution of a data use agreement. Interested investigators are encouraged to contact the corresponding author.
References
Carter AM. Placental oxygen consumption. Part I: in vivo studies–a review. Placenta. 2000;21:S31–7.
Redline RW. Placental pathology: a systematic approach with clinical correlations. Placenta. 2008;29:86–91.
Khong TY, Mooney EE, Ariel I, Balmus NCM, Boyd TK, Brundler M-A, et al. Sampling and Definitions of Placental Lesions: Amsterdam Placental Workshop Group Consensus Statement. Arch Pathol Lab Med. 2016;140:698–713.
Catov JM, Muldoon MF, Reis SE, Ness RB, Nguyen LN, Yamal J-M, et al. Preterm birth with placental evidence of malperfusion is associated with cardiovascular risk factors after pregnancy: a prospective cohort study. BJOG. 2018;125:1009–17.
Mestan KK, Check J, Minturn L, Yallapragada S, Farrow KN, Liu X, et al. Placental pathologic changes of maternal vascular underperfusion in bronchopulmonary dysplasia and pulmonary hypertension. Placenta. 2014;35:570–4.
Blair E, de Groot J, Nelson KB. Placental infarction identified by macroscopic examination and risk of cerebral palsy in infants at 35 weeks of gestational age and over. Am J Obstet Gynecol. 2011;205:e1–7.
Barker DJP, Eriksson JG, Forsén T, Osmond C. Fetal origins of adult disease: strength of effects and biological basis. Int J Epidemiol. 2002;31:1235–9.
Kumar R, Yu Y, Story RE, Pongracic JA, Gupta R, Pearson C, et al. Prematurity, chorioamnionitis, and the development of recurrent wheezing: a prospective birth cohort study. J Allergy Clin Immunol. 2008;121:878–84.e6.
Roberts DJ. Placental pathology, a survival guide. Arch Pathol Lab Med. 2008;132:641–51.
Sun C-CJ, Revell VO, Belli AJ, Viscardi RM. Discrepancy in pathologic diagnosis of placental lesions. Arch Pathol Lab Med. 2002;126:706–9.
Cooper LAD, Carter AB, Farris AB, Wang F, Kong J, Gutman DA, et al. Digital pathology: data-Intensive Frontier in medical imaging: health-information sharing, specifically of digital pathology, is the subject of this paper which discusses how sharing the rich images in pathology can stretch the capabilities of all otherwise well-practiced disciplines. Proc IEEE Inst Electr Electron Eng. 2012;100:991–1003.
Jackson MR, Mayhew TM, Boyd PA. Quantitative description of the elaboration and maturation of villi from 10 weeks of gestation to term. Placenta. 1992;13:357–70.
Jauniaux E, Burton GJ. Pathophysiology of placenta accreta spectrum disorders: a review of current findings. Clin Obstet Gynecol. 2018;61:743–54.
Mukherjee R. Morphometric evaluation of preeclamptic placenta using light microscopic images. Biomed Res Int. 2014;2014:293690.
Al-Adnani M, Marnerides A, George S, Nasir A, Weber MA. “Delayed Villous Maturation” in placental reporting: concordance among consultant pediatric pathologists at a single specialist center. Pediatr Dev Pathol. 2015;18:375–9.
Turowski G, Vogel M. Re-view and view on maturation disorders in the placenta. APMIS. 2018;126:602–12.
Grether E, Redline B, Benirschke N. Reliability of placental histology using archived specimens. Paediatr Perinat Epidemiol. 1999;13:489–95.
Manuck TA, Rice MM, Bailit JL, Grobman WA, Reddy UM, Wapner RJ, et al. Preterm neonatal morbidity and mortality by gestational age: a contemporary cohort. Am J Obstet Gynecol. 2016;215:103.e1–103.e14.
Kalish RB, Chervenak FA. Sonographic determination of gestational age. Ultrasound Rev Obstet Gynecol. 2005;5:254–8.
Kalish RB, Thaler HT, Chasen ST, Gupta M, Berman SJ, Rosenwaks Z, et al. First- and second-trimester ultrasound assessment of gestational age. Am J Obstet Gynecol. 2004;191:975–8.
Taipale P. Predicting delivery date by ultrasound and last menstrual period in early gestation. Obstet. Gynecol. 2001;97:189–94.
Maly A, Goshen G, Sela J, Pinelis A, Stark M, Maly B. Histomorphometric study of placental villi vascular volume in toxemia and diabetes. Hum Pathol. 2005;36:1074–9.
Leavey K, Benton SJ, Grynspan D, Bainbridge SA, Morgen EK, Cox BJ. Gene markers of normal villous maturation and their expression in placentas with maturational pathology. Placenta. 2017;58:52–9.
Salsabili S, Mukherjee A, Ukwatta E, Chan ADC, Bainbridge S, Grynspan D. Automated segmentation of villi in histopathology images of placenta. Comput Biol Med. 2019;113:103420.
Swiderska-Chadaj Z, Markiewicz T, Koktysz R, Cierniak S. Image processing methods for the structural detection and gradation of placental villi. Comput Biol Med. 2018;100:259–69.
Ferlaino M, Glastonbury CA, Motta-Mejia C, Vatish M, Granne I, Kennedy S, et al. Towards deep cellular phenotyping in placental histology. Amsterdam, the Netherlands: The 1st Conference on Medical Imaging with Deep Learning, 2018.
Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velázquez Vega JE, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci USA. 2018;115:E2970–9.
Courtiol P, Maussion C, Moarii M, Pronier E, Pilcer S, Sefta M, et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat Med. 2019;25:1519–25.
Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25:1301–9.
Bychkov D, Linder N, Turkki R, Nordling S, Kovanen PE, Verrill C, et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep. 2018;8:3395.
Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181–93.e7.
Yan C, Nakane K, Wang X, Fu Y, Lu H, Fan X, et al. Automated gleason grading on prostate biopsy slides by statistical representations of homology profile. Comput Methods Programs Biomed. 2020;194:105528.
Shanes ED, Mithal LB, Otero S, Azad HA, Miller ES, Goldstein JA. Placental pathology in COVID-19. Am J Clin Pathol. 2020;154:23–32.
Freedman AA, Goldstein JA, Miller GE, Borders A, Keenan-Devlin L, Ernst LM. Seasonal variation of chronic villitis of unknown etiology. Pediatr Dev Pathol. 1093526619892353 (2019)
Macenko M, Niethammer M, Marron JS, Borland D, Woosley JT, Xiaojun Guan, et al. A method for normalizing histology slides for quantitative analysis. In 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA: IEEE; 2009. pp. 1107–1110.
Simard PY, Steinkraus D, Platt JC. Best practices for convolutional neural networks applied to visual document analysis. In Seventh International Conference on Document Analysis and Recognition, 2003. Proceedings., (Edinburgh, UK: IEEE Comput. Soc), 958–963 (2003).
Deng J, Dong W, Socher R, Li L-J, Kai Li, Li Fei-Fei ImageNet: A large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition, 2009. pp. 248–255.
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–58.
Christians JK, Grynspan D. Placental villous hypermaturation is associated with improved neonatal outcomes. Placenta. 2019;76:1–5.
Pinar H, Sung CJ, Oyer CE, Singer DB. Reference values for singleton and twin placental weights. Pediatr Pathol Lab Med. 1996;16:901–7.
Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst, Man, Cybern. 1979;9:62–6.
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int J Comput Vis. 2020;128:336–59.
Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell. 2019;1:206–215.
Clymer D, Kostadinov S, Catov J, Skvarca L, Pantanowitz L, Cagan J, et al. Decidual vasculopathy identification in whole slide images using multiresolution hierarchical convolutional neural networks. Am J Pathol. 2020;190:2111–22.
Hartman D, Roy S, Pantanowitz L, Amin M, Seethala R, Ishtiaque A, et al. Smartphone adapters for digital photomicrography. J Pathol Inform. 2014;5:24.
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–401.
Yuan Y. Spatial heterogeneity in the tumor microenvironment. Cold Spring Harb Perspect Med. 2016;6:a026583.
Zilenaite D, Rasmusson A, Augulis R, Besusparis J, Laurinaviciene A, Plancoulaine B, et al. Independent prognostic value of intratumoral heterogeneity and immune response features by automated digital immunohistochemistry analysis in early hormone receptor-positive breast carcinoma. Front Oncol. 2020;10:950.
Larsen BT, Smith ML, Elicker BM, Fernandez JM, de Morvil GAA-O, Pereira CAC, et al. Diagnostic approach to advanced fibrotic interstitial lung disease: bringing together clinical, radiologic, and histologic clues. Arch Pathol Lab Med. 2017;141:901–15.
Acknowledgements
The authors wish to acknowledge the efforts of clinical pathologists, residents, and staff in producing the original slides and reports.
Funding
JAG was supported by funding from the National Institutes of Health National Institute of Biomedical Imaging and Bioengineering (NIBIB K08EB030120). Slide scanning was supported by the Northwestern University Pathology Core Facility and a Cancer Center Support Grant (NCI CA060553). PM and LADC were supported by funding from the National Institutes of Health National Cancer Institute Grants (NCI U24CA19436201 and NCI U01CA220401).
Author information
Authors and Affiliations
Contributions
JAG and LADC conceived of the work. JAG, PM, and LADC developed methods. JAG prepared histologic slides and data. PM developed the model and performed the machine learning. JAG, PM, and LADC analyzed the results. PM and JAG generated figures. JAG, PM, and LADC wrote the paper. JAG takes responsibility for the integrity of the data and the accuracy of the data analysis.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by the Northwestern University Institutional Review Board (STU00211333)
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Mobadersany, P., Cooper, L.A.D. & Goldstein, J.A. GestAltNet: aggregation and attention to improve deep learning of gestational age from placental whole-slide images. Lab Invest 101, 942–951 (2021). https://doi.org/10.1038/s41374-021-00579-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00579-5
This article is cited by
-
Mapping cell-to-tissue graphs across human placenta histology whole slide images using deep learning with HAPPY
Nature Communications (2024)
-
Efficient diagnosis of IDH-mutant gliomas: 1p/19qNET assesses 1p/19q codeletion status using weakly-supervised learning
npj Precision Oncology (2023)
-
A Patch-Based Deep Learning Approach for Detecting Rib Fractures on Frontal Radiographs in Young Children
Journal of Digital Imaging (2023)
-
Genetic mutation and biological pathway prediction based on whole slide images in breast carcinoma using deep learning
npj Precision Oncology (2021)