Abstract
Rett syndrome (RTT) is a rare neurodevelopmental disorder that results in multiple disabilities. Exosomal microRNA (miRs) from urine-derived stem cells (USCs) have been shown to induce neurogenesis and aid in functional recovery from brain ischemia. In the present study, we sought to determine whether that exosomal miR-21-5p from USCs could promote early neural formation in a model of RTT. USCs were isolated and evaluated by flow cytometry. Exosomes were analyzed by transmission electron microscopy, tunable resistive pulse sensing (TRPS), and western blotting. PKH26 fluorescent dyes were used to observe intake of exosomes in vivo and in vitro. An RTT mouse model was treated with exosomes for behavioral studies. Dual‐luciferase report gene assays were conducted to evaluate the relationship between miR-21-5p and Eph receptor A4 (EphA4). In vitro, treatment with exosomes from human urine‐derived stem cells (USC-Exos) increased the percentage of neuron-specific class III beta-tubulin (Tuj1)+ nerve cells as well as the transcription levels of β-III tubulin and doublecortin (DCX). A higher level of miR-21-5p was observed in USC-Exos, which promoted differentiation in NSCs by targeting the EPha4/TEK axis. In vivo, exosomal miR-21-5p improved the behavior, motor coordination, and cognitive ability of mice, facilitated the differentiation of NSCs in the subventricular zone of the lateral ventricle and promoted a marked rise in the number of DCX+ cells. Our data provide evidence that exosomal miR-21-5p from human USCs facilitate early nerve formation by regulating the EPha4/TEK axis.
Similar content being viewed by others
Introduction
Rett syndrome (RTT) is a rare, progressive neurodevelopmental disease that mainly affects females (1 in 10,000 live births), with a period of 6 months of normal neurodevelopment [1]. It is reported that patients with RTT show symptoms of early neurologic deterioration at onset and a gradual loss of acquired cognitive, social, and motor skills [2]. RTT is an X-linked, neurogenetic disorder caused by mutations in methyl-CpG-binding protein 2 (MECP2) gene and is a key cause of severe mental retardation in women [3]. MeCP2 regulates the expression of numerous downstream targets, including microRNAs (miRNAs or miRs) [4].
Recent studies have elucidated the importance of miRNAs in prenatal and adult neurogenesis, brain maturation, and synaptic plasticity [5,6,7]. For example, miR-21 exerts protective roles in Alzheimer’s disease, which might be dependent on the PDCD4/PI3K/AKT/GSK-3β pathway in vitro [8]. These observations reveal important roles of miRNA during neurogenesis. Exosomes are effective transmitters of intercellular information. The cargo of exosomes are proteins, lipids, mRNAs, and miRNAs which are transported to other cells to influence their functions and physiological states [9, 10]. It has been reported that exosomal miR-26a from urine-derived stem cells (USCs) induces neurogenesis as well as functional recovery from brain ischemia [11]. The Eph receptor A4 (EphA4) is the target gene of miR-21-5p. The regulatory role of EphA4 in neurogenesis has recently been described [12]. One study showed that EphA4 is inversely correlated with TEK receptor tyrosine kinase (TEK or Tie2) receptor signaling and EphA4–TEK receptor interaction is crucial for the remodeling of the pial collateral after stroke [13]. In the present study, we sought to determine the effects of exosomes on nerve function in an animal model of RTT and explore the involvement of USC-Exo on the Eph receptor in the central nervous system. Here we report the effect of exosomes from human urine‐derived stem cells (USC-Exos) on neural stem cells (NSCs) and elucidate the role of USC-Exosome expressing miR-21-5p and its downstream molecular regulatory mechanisms in RTT.
Materials and methods
Isolation and culture of human USCs
This study was conducted in line with the principiums of the Helsinki Declaration and was authorized by the ethics committee of The Second Hospital of Jilin University. Urine samples were obtained from six healthy volunteers with a median age of 25 years and the latest urine samples (200 mL) were added with 100 U/mL penicillin and 100 mg/mL streptomycin to prevent microbial contamination. Written informed consent was acquired from all participants.
The supernatant was removed after centrifugation at 400×g for 10 min. Then, the cell pellets were resuspended in Dulbecco’s modified Eagle medium (DMEM). The cell suspension was transferred into gelatin-coated 24-well culture plates and then were cultured at 37 °C in a moist atmosphere with 5% carbon dioxide (CO2). After 7 days of culture, non-adherent cells were discarded and the medium was renewed. Colonies obtained from individual cells were labeled. Cells were subcultured upon reaching 80% confluence.
Flow cytometry
To block non-specific antigen binding, USCs at the fourth passage were cultured with 3% bovine serum albumin (BSA) for 30 min and were incubated with the monoclonal antibodies as follows (Abcam, Cambridge, UK): CD29-fluorescein isothiocyanate (FITC; ab21845), CD73-FITC (ab239246), CD90-FITC (ab11155), CD44-FITC (ab27285), CD45-FITC (ab27287), CD34-FITC (ab78165), and human leukemic D-related antigen (HLA-DR)-FITC (ab1182). Nonspecific fluorescence was detected by incubating similar cell aliquots with isotype-matched monoclonal antibodies (BD Biosciences, Sparks, MD). The antigens were analyzed by Guava easyCyte™ (MilliPore, Billerica, MA).
Multi-directional differentiation of USCs
Osteogenic differentiation
USCs at the fourth passage were cultured with osteogenic induction medium (Gibco, Invitrogen, Carlsbad, CA) when they grew to 80% confluence. After 21 days of induction, the osteogenic medium was discarded, and the cells were fixed with 4% paraformaldehyde, stained with alizarin red (Sigma-Aldrich, St. Louis, MO), and observed under microscope.
Adipogenic differentiation
USCs at the fourth passage were cultured with adipogenic induction medium (Gibco). The medium was changed every 3 days. After induction of 14 days, cells were fixed for 30 min and stained with Oil Red O stain for 10 min.
Chondrogenic differentiation
About 1 × 106 cells were centrifuged and added with chondrogenic medium (Gibco). After induction of 4 weeks, the cell slides were taken out, and the cultured cells were fixed in 4% paraformaldehyde solution at room temperature for 30 min, then stained with alcian blue for 30 min, dehydrated, cleared, and sealed with resin. All marked cells were identified by using an optical microscope (Leica, Germany).
Isolation and purification of USC-Exos
USCs were cultured with medium supplemented with exosome-removed FBS at 37 °C in 5% CO2 for 48 h. USCs-conditioned medium was centrifuged at 2500 × g at 4 °C for 20 min. The supernatant was then filtrated with 0.22-μm filters (MilliPore) to eliminate the cell debris. Then, the filtrated supernatant was transferred into ultra-clear tube (MilliPore) and ultracentrifuged at 100,000 × g at 4 °C for 70 min to settle the USC-Exos. Next, USC-Exos were resuspended in 200 μL of sterile PBS. For further purification, the solution containing USC-Exos was added to a sterile Ultra-Clear™ tube (Beckman Coulter, Kraemer Boulevard Brea, CA) containing 30% sucrose/D2O cushion and ultracentrifuged at 100,000 × g at 4 °C for 70 min (Beckman Coulter, Sorvall, Avanti J-26XP, fixed angle rotor). USC-Exos were recovered. The protein content of USC-Exos was detected with the BCA Protein Assay Kit (Thermo Fisher Scientific, Austin, Texas). The absorbance was read at 562 nm by using a microplate reader (Bio-Rad Laboratories, California).
Transmission electron microscopy
USC-Exos were fixed in 3% (w/v) glutaraldehyde and 2% PFA buffer, and then were placed on copper grids coated with Formvar. After washing, the grids were contrasted in 2% uranyl acetate, dried, and were checked with the transmission electron microscope (Morgagni 268D, Philips, Holland).
Tunable resistive pulse sensing
Tunable resistive pulse sensing (TRPS) was conducted to assess concentration and size distribution of USC-Exos, which was implemented using the qNano platform with an NP100-rated nanopore (Izon Science, Oxford, UK). The membrane was extended at 43.0 mm. CPC100 corpuscules (Izon Science) were applied for the calibration of concentration and size according to manufacturer’s descriptions. USC-Exos samples were diluted and measured for 3 min. Data processing and analysis were conducted using Izon Control Suite software v2.2 (Izon Science).
Uptake of USC‐exos
PKH26 Red Fluorescent Cell Linker kits (Sigma-Aldrich) were used to label exosomes. Briefly, 2 μM PKH26 was added to the exosomal suspension and incubated under sterile conditions for 4 min. After adding PBS containing 1% BSA, the exosomal suspension was centrifuged at 135,000 × g for 1 h at 4 °C in Beckman TLA110 and resuspended in PBS. And 2 μL of exosomes labeled with PKH26 were implanted into the lateral ventricle (LV) of each cerebral hemisphere of mice. After 12 h, the brain tissue sections were determined with fluorescence microscope. In vitro, PKH26-labeled exosomes were incubated with NSCs for 4 h and were observed using the fluorescence microscope (Leica).
RTT mice model
Experiments were carried out using the B6.129S-MeCP2tm1Hzo mouse model for RTT (MeCP2308 truncation model). Wild-type (WT) B6 male mice and heterozygous female mice were bred to obtain MeCP2 hemizygous male mice and their WT offspring. Grown heterozygous female mice were bought from Jackson Laboratory. Because the inactivation of X chromosome in female mice leads to unpredictable phenotypes, MeCP2 hemizygous male mice and their WT (5 months old) were used to replace heterozygous female mice. Animals were kept in specific pathogen-free environments and maintained a 12-h light/dark cycle. All procedures and experiments were permitted by the Animal Experiment Ethics Committee (ZJU20160281) [14].
Exosomal therapy
The experiments in vivo were divided into six groups (10 mice per group), including (1) WT mice; (2) RTT mice; (3) RTT mice treated with PBS, which were injected with 2 μL of PBS into the LV of each cerebral hemisphere; (4) RTT mice treated with USC-Exos, which were injected with 2 μL of USC-Exos into the LV of each cerebral hemisphere; (5) RTT mice treated with NC-exo, which were injected with 2 μL of NC-exo into the LV of each cerebral hemisphere; (6) RTT mice treated with inhibitor-exo, which were injected with 2 μL of inhibitor-exo into the LV of each cerebral hemisphere (all every other day for 3 weeks). The injection was performed manually with a 1.0 mm glass needle (World Precision Instruments) pulled out by the P-97 flame/brown micropipette extractor (Sutter Instruments), and a glass needle with a graduated 1 μL volume was connected to the syringe. Behavioral researches were performed after the 21st injection and the follow-up 4-h washout period. The mice were immolated by cervical luxation, the frontal cortex was then analyzed and quickly frozen for biochemical analysis.
Behavioral research
Behavior
Mice were kept in cages, enclosed by cardboard boxes with barbed wire and joined to the edge of a coop. At the blocked end of the tube outside the cage, the mice were provided with ground peanuts and the cage was supplied with water and food. After a certain amount of time, the content of nut meal in the coop and the residual amount in the silk drum were measured to detect food hoarding behavior of mice. At 20 min, six pieces of 4 cm × 4 cm square paper were set in the left upper corner of the cage and nesting score was performed at 8 o’clock on the following day (Table 1).
Movement and coordination
Mice were hooked with 10 grades of wire balls weighing from 100 g to 190 g, respectively. The experimenter raised the mid of the mouse’s tail and determined each mouse’s grip force on the steel ball for 3 s per level, which started at a weight level of 100 g. The average hanging time and distance of the three tests were calculated. The conversion point of the rope grab test result was as follows: average suspension time C 10 × average distance. The 160 cm iron rod with a diameter of 0.6 cm was set on the white iron basin. The movement of mice was observed for 60 s and then we measured the time for each mouse to maintain balance, which was repeated three times (Table 2).
Cognitive function
A swimming pool was separated into four quadrants on average. A circular concealed platform was set in the middle of the target quadrant and kept unaltered throughout the experiment. The mice were set in the water, and the time for the mice to find and board the platform was timed and referred to as the escape latency. In case the mouse could not find the platform within 2 min, the experimenter rescued the mouse. In this instance, the escape delay was recorded as 2 min. On the fifth day of the water maze experiment, the platform was dismantled, and mice were set in the water. Finally, the swimming path was timed for 2 min, and the results were analyzed statistically. The swimming path score scale is shown in Table 3.
Brain tissue preparation and immunofluorescence
After the behavioral studies, mice were euthanized by cervical luxation. The brain was taken out, fixed in 4% PFA at 4 °C overnight and soaked in 30% sucrose for 3 days for cryoprotection. After burying the brain in a compound with optimal cutting temperature and freezing, the brain tissues (20 μm) were stained with doublecortin (DCX, 1:100, Cell Signaling Technology, Beverly, MA) specific markers and images were obtained with a Leica microscope.
Isolation and enrichment of mouse NSCs
Mouse NSCs were isolated from embryonic brain tissues of WT mice and RTT mice. In short, the cerebral cortex tissue was isolated from the embryo on day 13.5 (E13.5) and subjected to physical grinding. The separated tissues were filtered through a 40-μm filter, and single cells were cultured in a matrix-free tissue culture flask to form neurospheres in NSCs proliferation medium. Primary neurospheres were gathered, separated into single cells with Accuttase (Sigma-Aldrich) for 5 min and reformed. After three times of neurosphere formations, abundant NSCs were obtained for immunocytochemical analysis.
Differentiation of NSCs
About 5 × 104 NSCs were planted in a 24-well plate of poly-l-ornithine/laminin-coated coverslips and cultured with DMEM/F12 medium for 1–2 weeks.
Immunocytochemistry
The cultured cells were fixed in 4% PFA for 20 min, washed using PBS for three times, penetrated with 0.2% Triton X-100 in PBS for 10 min, blocked with 2% BSA for 1 h, and incubated with primary antibodies mouse anti-neuron-specific class III beta-tubulin (Tuj1; Covance, Princetown, NJ, 1:500), Nestin (Abcam, Ab6142, 1:100), and F-actin (Abcam, ab205, 1:100) at 4 °C overnight. After washed and incubated for 1 h at room temperature, the cells were added and incubated with secondary goat anti-mouse immunoglobulin G (IgG; Abcam, ab150113, 1:500 or ab150115, 1:500). Nuclear DNA was marked with 4,6-diamidino-2-phenylindole (DAPI). Nuclear DNA was marked with 4,6-diamidino-2-phenylindole (DAPI). Then, immunostaining was examined by a Zeiss 710 confocal laser scanning microscope.
Transfection with electroporation
EPhA4 and TEK overexpressed plasmids and their controls, miR-21-5p inhibitor and their controls were purchased from RiboBio. FAM-miR-21-5p was purchased from GenePharma (Shanghai, China). The above plasmids were introduced into cells using the NucleofectorTM kit (Amaxa, Germany). Total RNA or protein was extracted 24 or 72 h after nuclear infection.
qRT-PCR
Total RNA was extracted and reversely transcribed by PrimeScriPt RT reagent Kit (RR047A, Takara, Japan). The complementary DNA was measured using Fast SYBR Green PCR kit (Applied Biosystems, Foster City) and StepOnePlus Real-Time PCR System (Applied Biosystems). For miRNA, mirVana qRT-PCR miRNA Detection kit (Thermo Fisher Scientific) was used for detection. Each hole was set for three repetitions. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 served the internal reference for mRNA and miRNA, respectively. To analyze the related expression of genes and miRNA, the 2−ΔΔCt method was adopted. The primer design is shown in the Table 4.
Western blots
The cells or exosomes were lysed with Radio-Immunoprecipitation Assay lysate (MilliPore). The separated proteins by sodium dodecyl sulfate polyacrylamide gel electrophoresis were electrotransferred to polyvinylidene fluoride membranes, blocked with 5% BSA, and incubated with diluted primary antibodies at 4 °C overnight respectively, including rabbit anti-CD9 (Abcam, ab92726, 1:2000), anti-CD63 (Abcam, ab59479, 1:1000), anti-CD81 (Abcam, ab109201, 1:1000), calnexin (Abcam, Ab22595, 1:1000), EPha4 (Abcam, ab5396, 1:1000), anti-Alix (Abcam, ab186429, 1:1000), and GAPDH (Abcam, ab181602, 1:10,000). After washing, the membrane was incubated with horseradish peroxidase (HRP)-labeled goat anti-rabbit secondary antibody (Abcam, ab205719, 1:2000). Electrogenerated chemiluminescence fluorescence detection kit (Cat. No. BB- 3501, Amersham, Little Chalfont, UK) was used for exposure imaging. Bio-Rad image analysis system (Bio-Rad Laboratories) and Quantity One v4.6.2 software were applied to analyze the expression of proteins.
Dual luciferase reporter assay
Predicted target binding sites were obtained from Targetscan. The synthetic EPha4 3ʹ-untranslated regions (3ʹ-UTRs) of the WT and mutant (MUT) were plugged into the luciferase reporter vector PmiR-RB-RePort™ (product number: VT1399, U-Bio, Shanghai, China). The correctly sequenced luciferase reporter plasmids WT and MUT were co-transfected into HEK293 cells with miR-21-5p mimic or mimic NC. After 48 h, cells were lysed and the luciferase activity was identified using a luciferase detection kit (K801-200, Biovision, San Francisco) and a Glomax 20/20 luminometer fluorescence detector (Promega, Madison, WI).
Statistical analysis
The statistical software SPSS 21.0 was used. Measurement data were shown as mean ± standard deviation. Unpaired t test and single factor analysis of variance were applied for the comparison between several groups. Tukey’s post-hoc test was conducted. P < 0.05 means the statistically significant difference.
Results
Identification of USCs and USC-Exos
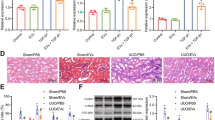
USCs were able to differentiate into neuronal cells. After being transplanted into rat brains, USCs survived, migrated, and differentiated into neuronal lineages at the injury site, which makes USCs a more promising choice than mesenchymal stem cells (MSCs) [11]. USCs were isolated from fresh urine. USCs were confirmed to have fibroblast-like morphology using optical microscopy (Fig. 1A) and reached 80–90% confluence after 16 days (Fig. 1B). After several passages, USCs maintained a long spindle shape (Fig. 1C). When cultured in osteogenic, adipogenic, or chondrogenic medium, USCs could differentiate into osteoblasts, adipocytes, and chondrocytes (Fig. 1D–F). Therefore, the features of USCs were in line with the standard for defining multipotent MSCs.
Morphology and growth of USCs, scale bar: 50 µm (A–C). USCs differentiate into osteoblasts by alizarin red staining (D), adipocytes by Oil Red O staining (E), chondrocytes by alcian blue staining (F), scale bar: 25 µm. The morphology of USC-Exo was observed under TEM, scale bar: 100 nm (G). The size of USC-Exo was detected by TRPS and the measured average concentration (particles/mL) was 5.1E+004 (H). Exo-specific proteins CD9, CD63, Alix, CD81, and endoplasmic reticulum-specific protein calnexin were detected by western blot (I).
USC-Exos were reported to promote neurogenesis after ischemic stroke [11]. To explore the role of USC-Exos in RTT, we extracted and identified USC-Exos. The morphology and size of USC-Exos were observed by transmission electron microscopy (TEM) and Nano Sight, respectively. The TEM results showed that USC-Exos were a spherical vesicle at about 100 nm (Fig. 1G). As expected, TRPS analysis demonstrated that the particle size of USC-Exos was mainly distributed at 50–100 nm (Fig. 1H). Exosomal markers CD9, CD63, Alix, and CD81 were expressed in USC-Exos, but the endoplasmic reticulum-specific protein calnexin was not (Fig. 1I). The above results revealed that the isolated exosomes could be used in subsequent experiments.
USC-Exos promoted the differentiation of RTT NSCs
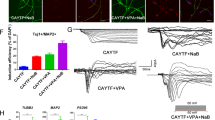
Several studies have reported the potential changes in the neuronal differentiation, migration, and branching of RTT ipsilateral neurons [15, 16]. We then isolated NSCs from embryonic brain tissues of WT mice and RTT308 mice. USCs cultured in vitro could form the free-floating neurospheres (Fig. 2A). Immunocytochemistry showed that most cells in the neurosphere were Nestin-positive cells (Fig. 2B), indicating that neurospheres had the phenotype of NSCs. To judge whether NSCs could internalize USC-Exos, USC-Exos were marked with PKH26 fluorescent dye and cultured with NSCs. After incubation for 4 h, USC-Exos labeled with PKH26 were effectively internalized by NSCs (Fig. 2C). The influences USC-Exos exerted on the differentiation of NSCs were measured by immunofluorescence. NSCs and exosomes (15 μg/mL) were co-cultured in differentiation medium and were labeled with Tuj1 antibody after 7 days of incubation. The results of immunofluorescence detection showed that compared with the normal NSCs group, percentage of Tuj1+ nerve cells in the RTT group decreased (Fig. 2D); compared with the RTT + PBS group, percentage of Tuj1+ nerve cells in the RTT + USC-Exos group was increased. Compared with the normal NSCs group, transcription levels of β-III tubulin and DCX in the RTT group were reduced; compared with the RTT + PBS group, transcription levels of β-III tubulin and DCX in the RTT + USC-Exos group were increased (Fig. 2E).
The morphology of NSCs was observed under a light microscope, scale bar: 50 µm (A). Immunocytochemistry showed that most cells in the neurosphere were Nestin-positive cells, scale bar: 25 µm (B). USC-Exos labeled with PKH26 was effectively internalized by NSCs, scale bar: 25 µm (C). Immunofluorescence staining and quantification of Tuj1 positive cells, scale bar: 25 µm (D, F). The mRNA expression levels of β-III tubulin and DCX in NSCs were determined by qRT-PCR (E, G). *P < 0.05 (The data results were measurement data and shown as mean ± standard deviation. Independent sample t test and one-way analysis of variance were, respectively, used for the comparison between two groups and several groups, with Tukey’s post-hoc test. The experiment was repeated three times).
Since cells communicate with each other by secreting soluble factors, we compared the influences that exosome-conditioned medium (CM) and exosome-free CM (supernatant after ultracentrifugation, SN) exerted on the differentiation of RTT NSCs. Immunocytochemistry and qPCR analysis showed that exosomes and CM had a similar effect on the differentiation of NSCs (Fig. 2F, G), while SN had no effect on differentiation of NSCs, suggesting that exosomes rather than soluble factors might regulate neurogenic microenvironment.
USC-Exos promoted neural differentiation of RTT NSCs by regulating EPha4/TEK axis
There was evidence that EPha4 was a potential regulator of neurogenesis and negatively regulated the formation of collateral pia mater arterioles [17,18,19,20]. We confirmed EPha4 expression in RTT NSCs (Fig. 3A). Compared with the normal group, EPha4 expression in the RTT group was increased. Compared with the RTT + PBS group, EPha4 expression in the RTT + USC-Exos group was downregulated. Further, EPha4 expression in NSCs, which were transfected with EPha4-overexpressed plasmid, was upregulated (Fig. 3B). Then, compared with the oe-NC group, percentage of Tuj1-postive nerve cells in the oe-NC + USC-Exos group increased; compared with the oe-NC + USC-Exos group, percentage of Tuj1-positive nerve cells in the oe-EPha4 + USC-Exos group decreased (Fig. 3C). The transcription levels of β-III tubulin and DCX in the oe-NC + USC-Exos group were notably higher than that in the oe-NC group; compared with the oe-NC + USC-Exos group, the transcription levels of β-III tubulin and DCX in the oe-EPha4 + USC-Exos group decreased (Fig. 3D). Compared with the oe-NC group, the protein levels of EPha4 in the oe-NC + USC-Exos group decreased; compared with the oe-NC + USC-Exos group, the protein levels of EPha4 in the oe-EPha4 + USC-Exos group were importantly enhanced (Fig. 3E).
EPha4 expression in NSCs of each group was detected by western blot (A, E). The transfection efficiency of EPha4 in NSCs was detected by western blot (B). qRT-PCR detected the mRNA expression of β-III tubulin and DCX (C). Immunofluorescence staining and quantification of Tuj1-positive cells (D, H). TEK protein expression in NSCs was detected by western blot (F). TEK transfection efficiency in NSCs by qRT-PCR (G). *P < 0.05 (The data results were measurement data and were shown as mean ± standard deviation. The comparison between the two groups was analyzed by independent sample t test. One-way analysis of variance was used for the comparison between several groups, with Tukey’s post-hoc test. The experiment was performed three times).
TEK has been reported to promote differentiation of NSCs and EPha4 is a negative regulator of TEK [13, 21]. Western blot results revealed that compared with the oe-NC group, TEK expression in the oe-EPha4 group decreased (Fig. 3F). In order to investigate whether EPha4 negatively regulated TEK receptors to inhibit NSC differentiation, we transfected RTT NSCs with plasmids that highly express TEK gene. The results of qRT-PCR showed that TEK expression in NSCs transfected with TEK overexpression plasmid was upregulated (Fig. 3G). The transcription levels of β-III tubulin, DCX, Nestin, and SOX2 in cells were analyzed. qRT-PCR analysis indicated that compared with the oe-NC group, the transcription levels of β-III tubulin, DCX, Nestin, and SOX2 in the oe-TEK group were importantly enhanced; compared with the oe-EPha4 + oe-NC group, the transcription levels of β-III tubulin, DCX, Nestin, and SOX2 in the oe-EPha4 + oe-TEK group were increased (Fig. 3H), indicating that EPha4 inhibited the differentiation of NSCs by negatively regulating TEK receptors. The above data indicated that USC-Exos-induced differentiation of RTT NSCs might be partially due to the inhibition of EPha4, which promoted the expression of TEK.
USC-Exos-mediated miR-21-5p promoted neuronal differentiation of RTT NSCs by targeting EPha4
In recent years, the emerging evdience has proved the importance of miRNAs in prenatal and adult neurogenesis, brain maturation, and synaptic plasticity [5,6,7]. Exosomes can mediate cell-to-cell communication by transferring miRNAs. Therefore, we predicted the upstream regulator of EPha4 by bioinformatics. The Targetscan website predicted that miR-21-5p and EPha4 had a complementary pairing in the 3ʹUTR region (Fig. 4A). We used dual luciferase activity assays to assess the binding capacity and designed wild-type EPha4 (WT-EPha4) and mutant (MUT-EPha4) sequences. The results of dual luciferase activity assays showed that the luciferase activity of WT-EPha4 in the miR-21-5p mimic group was lower than the mimic NC group, while there was no difference in the luciferase activity of MUT-EPha4 between the two groups (Fig. 4B). In addition, miR-21-5p had a higher expression in USC-Exos (Fig. 4C). To verify the role of miR-21-5p in USC-Exos-mediated NSCs differentiation, miR-21-5p inhibitors or controls were transfected into USCs (Fig. 4D), and miR-21-5p expression in exosomes (inhibitor-Exos or NC-Exos) isolated from these USCs was analyzed. Compared with NC-Exos, miR-21-5p expression in inhibitor-Exos was reduced (Fig. 4E). In addition, it was found that miR-21-5p expression in the USC-Exos treatment group was higher than that in the PBS group, and that in the NC-Exos group was importantly higher than the inhibitor-Exos group (Fig. 4F). Next, inhibitor-Exos or NC-Exos were incubated with RTT NSCs to explore the effect of miR-21-5p on EPha4 expression in RTT NSCs (Fig. 4G). Compared with NC-Exos, inhibitor-Exos could not inhibit EPha4 expression in RTT NSCs. Moreover, the effect of exogenous miR-21-5p on the differentiation of RTT neurons was also evaluated. Compared with NC-Exos, the differentiation of NSCs decreased after the treatment with inhibitor-Exos (Fig. 4H). Compared with the NC-Exos group, the transcription levels of β-III tubulin and DCX in the inhibitor-Exos group were reduced (Fig. 4I). Thus, miR-21-5p targeting EPha4 through exosome transfer was one of the mechanisms that promote neurogenesis.
The Targetscan website predicted that miR-21-5p and EPha4 had a complementary pairing in the 3ʹUTR region (A). Dual luciferase activity assays were used for evaluation of the binding ability of miR-21-5p to EPha4 (B). The expression of miR-21-5p in USCs and USC-Exos was detected by qRT-PCR (C). The transfection efficiency of miR-21-5p in USCs was measured by qRT-PCR (D). The expression of miR-21-5p in USC-Exos was detected by qRT-PCR (E). The expression of miR-21-5p expression in NSCs was detected by qRT-PCR (F). EPha4 expression in NSCs of each group was detected by western blot (G). Immunofluorescence staining of TUJ1 positive cells (H). The mRNA expression levels of β-III tubulin and DCX in NSCs were detected by qRT-PCR (I). *P < 0.05 (The data results were measurement data and shown as mean ± standard deviation. The comparison between the two groups was analyzed by independent sample t test. One-way analysis of variance was used for the comparison between multiple groups, with Tukey’s post-hoc test. The experiment was performed three times).
Exosomal miR-21-5p promoted neurogenesis in RTT mice
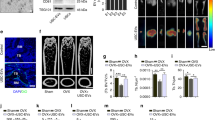
Then, we investigated the role of USC-Exos in the RTT308 mouse animal model. To ascertain whether the infused USC-Exos could move into the brain, USC-Exos were marked with PKH26 fluorescent dye and injected into mice. Fluorescence of PKH26 labeling in the brain of mice in the USC-Exos treatment group confirmed that USC-Exos could enter the brain (Fig. 5A). The subventricular zone (SVZ) of the LV was the main area of NSCs, which might be a latent therapeutic target area for USC-Exos. Compared with the WT group, SVZ DCX+ (early neuronal marker DCX) cells of RTT mice were importantly reduced; compared with the RTT + PBS group, SVZ DCX+ cells of RTT mice in the RTT + USC-Exos group increased; compared with the RTT + NC-Exos group, SVZ DCX+ cells of RTT mice in the RTT + inhibitor-Exos group were decreased. Next, we used qRT-PCR to detect miR-21-5p expression and DCX mRNA in the subventricular tissues of the LV (Fig. 5B). Compared with the WT group, miR-21-5p and DCX mRNA expression in the RTT group dropped. Compared with the RTT + PBS group, miR-21-5p and DCX mRNA expression in the RTT + USC-Exos group were importantly increased. Compared with the RTT + NC-Exos group, miR-21-5p and DCX mRNA expression in the RTT + inhibitor-Exos group were reduced (Fig. 5C). Compared with the WT group, EPha4 expression in the RTT group was notably increased; compared with the RTT + PBS group, EPha4 expression in the RTT + USC-Exos group reduced; compared with the RTT + NC-Exos group, EPha4 expression in the RTT + inhibitor-Exos group was increased (Fig. 5D). Exosomal miR-21-5p promoted RTT mice neurogenesis, which could help to improve the functional outcomes.
The USC-Exos were marked with PKH-26 fluorescent dye and implanted into the mice to observe the fluorescence intensity, scale bar: 25 µm (A). Representative immunofluorescence staining and quantitative analysis were used for DCX (green) positive cells in the subventricular zone of mice and DAPI was used to stain the nuclei, scale bar: 25 µm (B). The expression levels of miR-21-5p and DCX mRNA in the subventricular tissue of the lateral ventricle were detected by qRT-PCR (C). EPha4 protein expression in subventricular tissue of lateral ventricle was detected by western blot (D). *P < 0.05 (The data results were measurement data and were expressed as mean ± standard deviation. Independent sample t test was applied to analyze the comparison between two groups and there were ten mice in each group).
Exosomal miR-21-5p treatment improved the behavior, motor coordination function, and cognitive ability of RTT308 mice
The food storage capacity of animals in each group was shown in the figure (Fig. 6A). Compared with the WT group, the food storage capacity of RTT mice was notably reduced; compared with the RTT + PBS group, the food storage capacity of the RTT + USC-Exos group was enhanced; compared with the RTT + NC-Exos group, the food storage capacity of the RTT + inhibitor-Exos group was reduced. However, there was no notable difference in nesting ability between the mice in each group (Fig. 6B). The results of animal movement coordination in each group were shown in the figure (Fig. 6C). Compared with WT mice, the grabbing force and ability of RTT mice were importantly lower; compared with the RTT + PBS group, the grabbing force and ability of mice in the RTT + USC-Exos group were enhanced; compared with the RTT + NC-Exos group, the grabbing force and ability of the mice in the RTT + inhibitor-Exos group were importantly reduced. There was no notable difference in the balance beam test among mice in each group (Fig. 6D). The Morris water maze experiment (Fig. 6E) showed that compared with WT mice, the time for RTT mice to find the hidden platform was notably increased; compared with the RTT + PBS group, the time to find the hidden platform in the RTT + USC-Exos group was shortened. Compared with the RTT + NC-Exos group, the time for the RTT + inhibitor-Exos group to find the hidden platform increased. There was no important difference in the spatial search capability of mice in each group (Fig. 6F). Thus, the behavior, motor coordination, and cognitive function of RTT mice were notably reduced, and treatment with human USC-derived exosomal miR-21-5p could improve these behaviors.
Food storage capacity of mice in each group (A). Nesting capability of mice in each group (B). The grabbing force and ability of the mice in each group (C). Balance beam test of mice in each group (D). The time when the mice in each group discovered the hidden platform (E). Spatial learning ability of mice in each group (F). *P < 0.05 (The data results were measurement data and were expressed as mean ± standard deviation. The comparison between the two groups was analyzed by independent sample t test and there were ten mice in each group).
Discussion
Exosomes possess various roles in the nervous system and exert pivotal functions in neurodegenerative diseases and synaptic pruning [22]. There is evidence that USCs differentiate into neural lineage cells and that their multi-lineage differentiation potentials may be an attractive cell-based therapy in neurology [23]. Furthermore, USCs-derived exosomes promote not only proliferation but also neuronal differentiation of NSCs after oxygen–glucose deprivation/reoxygenation [11]. In the present study, we focused on the regulatory role of exosomal miRNAs from USCs in a model of RTT. After in vitro and in vivo assays, we concluded that exosomal miR-21-5p from USCs could promote early neurogenesis of RTT by regulating the EPha4-dependent TEK expression (Fig. 7).
USCs can be collected by noninvasive approaches and are easy to culture and differentiate, and show great potential as a continual source of exosomes [24]. Exosome-based therapy is a promising method of tissue engineering, because of their high stability, their lack of immune rejection, their non-tumorigenicity, and the fact that they do not produce vascular obstructions [25, 26]. Previously, USCs-Exo were shown to promote wound healing [27], induce angiogenesis, relieve lower limb ischemia, and suppress diabetic nephropathy [28, 29]. In this study, USC-Exo promoted differentiation of NSCs in a model of RTT. Moreover, since cells can communicate with each other by secreting soluble factors, we compared the effects of exosomes CM and exosome exclusion medium on the differentiation of NSCs in a model of RTT. Consequently, exosomes and CM have similar effects on NSC differentiation.
Eph receptors possess the functions in controlling cell proliferation, migration, boundary formation, and repulsive/attractive cues [30]. It is reported that EphA4 receptor can regulate the pial arteriole collateral formation in the brain [17]. What is more, EphA4 receptors regulate axonal guidance and fasciculation, neural crest migration, midline development, and synaptic plasticity within the central nervous system [31,32,33]. On the basis of these reports, we sought to understand the molecular mechanisms that regulate neuronal differentiation in RTT. First of all, we confirmed that EphA4 was highly expressed in NSCs of RTT and the addition of USC-Exos could inhibit the expression of EphA4. Second, we also demonstrated that EPha4 inhibited NSC differentiation by negatively regulating TEK.
Additionally, the differentiation of NSCs in RTT induced by USC-Exos may be partly attributed to the inhibition of EPha4. TEK, the cognate receptor for Ang1 and Ang2, is mainly expressed on the vascular endothelium and possess the potentials on cell survival, vascular leakage, and inflammatory genes [34]. It is reported that upregulated TEK can be observed in glioblastoma after radiotherapy [35]. However, previous evidence elaborated the important role of EphA4/TEK in functional deficits and neural tissue damage after permanent middle cerebral artery occlusion [13].
Recent reports have presented compelling arguments for the importance of miRNAs in prenatal and adult neurogenesis, brain maturation, and synaptic plasticity [5,6,7]. Exosomes can mediate cell–cell communication by transferring miRNAs [36]. Our results show that miR-21-5p is the upstream regulator of EphA4, and targeting EPha4 by miR-21-5p through exosome transfer was one of the mechanisms promoting neurogenesis. We also concluded that exosomal miR-21-5p promoted neurogenesis in mice with RTT. Interestingly, RTT mice showed reduced behavior, motor coordination, and cognitive function, which were improved by the exosomal miR-21-5p from USCs. These results indicate that the increase of miR-21-5p level in the brain contributes to improvement in neurological outcome [37]. Similarly, previous study also revealed that miR-145 could protect the viability of human cerebral cortical neurons after oxygen–glucose deprivation by decreasing EPHA4 [38].
In conclusion, this study shows that exosomal miR-21-5p from USCs promotes early neurogenesis by regulating EphA4-dependent TEK to induce NSC differentiation in a model of RTT, highlighting a potential USC-derived exosomes-based treatment for RTT.
References
Lyst MJ, Bird A. Rett syndrome: a complex disorder with simple roots. Nat Rev Genet. 2015;16:261–75.
Percy AK, Neul JL, Glaze DG, Motil KJ, Skinner SA, Khwaja O, et al. Rett syndrome diagnostic criteria: lessons from the Natural History Study. Ann Neurol. 2010;68:951–5.
Shah RR, Bird AP. MeCP2 mutations: progress towards understanding and treating Rett syndrome. Genome Med. 2017;9:17.
Castro J, Mellios N, Sur M. Mechanisms and therapeutic challenges in autism spectrum disorders: insights from Rett syndrome. Curr Opin Neurol. 2013;26:154–9.
Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr Opin Neurobiol. 2009;19:461–70.
Mellios N, Sur M. The emerging role of microRNAs in schizophrenia and autism spectrum disorders. Front Psychiatry. 2012;3:39.
Mellios N, Sugihara H, Castro J, Banerjee A, Le C, Kumar A, et al. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat Neurosci. 2011;14:1240–2.
Feng MG, Liu CF, Chen L, Feng WB, Liu M, Hai H, et al. MiR-21 attenuates apoptosis-triggered by amyloid-beta via modulating PDCD4/ PI3K/AKT/GSK-3beta pathway in SH-SY5Y cells. Biomed Pharmacother. 2018;101:1003–07.
Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15.
Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–25.
Ling X, Zhang G, Xia Y, Zhu Q, Zhang J, Li Q, et al. Exosomes from human urine-derived stem cells enhanced neurogenesis via miR-26a/HDAC6 axis after ischaemic stroke. J Cell Mol Med. 2020;24:640–54.
Khodosevich K, Watanabe Y, Monyer H. EphA4 preserves postnatal and adult neural stem cells in an undifferentiated state in vivo. J Cell Sci. 2011;124:1268–79.
Okyere B, Mills WA 3rd, Wang X, Chen M, Chen J, Hazy A, et al. EphA4/Tie2 crosstalk regulates leptomeningeal collateral remodeling following ischemic stroke. J Clin Invest. 2020;130:1024–35.
Yuan ZF, Mao SS, Shen J, Jiang LH, Xu L, Xu JL, et al. Insulin-like growth factor-1 down-regulates the phosphorylation of FXYD1 and rescues behavioral deficits in a mouse model of Rett syndrome. Front Neurosci. 2020;14:20.
Williams EC, Zhong X, Mohamed A, Li R, Liu Y, Dong Q, et al. Mutant astrocytes differentiated from Rett syndrome patients-specific iPSCs have adverse effects on wild-type neurons. Hum Mol Genet. 2014;23:2968–80.
Fernandes TG, Duarte ST, Ghazvini M, Gaspar C, Santos DC, Porteira AR, et al. Neural commitment of human pluripotent stem cells under defined conditions recapitulates neural development and generates patient-specific neural cells. Biotechnol J. 2015;10:1578–88.
Okyere B, Giridhar K, Hazy A, Chen M, Keimig D, Bielitz RC, et al. Endothelial-specific EphA4 negatively regulates native pial collateral formation and re-perfusion following hindlimb ischemia. PLoS ONE. 2016;11:e0159930.
Adams RH, Wilkinson GA, Weiss C, Diella F, Gale NW, Deutsch U, et al. Roles of ephrinB ligands and EphB receptors in cardiovascular development: demarcation of arterial/venous domains, vascular morphogenesis, and sprouting angiogenesis. Genes Dev. 1999;13:295–306.
Canty AJ, Greferath U, Turnley AM, Murphy M. Eph tyrosine kinase receptor EphA4 is required for the topographic mapping of the corticospinal tract. Proc Natl Acad Sci USA. 2006;103:15629–34.
Gale NW, Holland SJ, Valenzuela DM, Flenniken A, Pan L, Ryan TE, et al. Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron. 1996;17:9–19.
Berlucchi M, Maroldi R, Aga A, Grazzani L, Padoan R. Ethmoid mucocele: a new feature of primary ciliary dyskinesia. Pediatr Pulmonol. 2010;45:197–201.
Sharma P, Mesci P, Carromeu C, McClatchy DR, Schiapparelli L, Yates JR 3rd, et al. Exosomes regulate neurogenesis and circuit assembly. Proc Natl Acad Sci USA. 2019;116:16086–94.
Guan JJ, Niu X, Gong FX, Hu B, Guo SC, Lou YL, et al. Biological characteristics of human-urine-derived stem cells: potential for cell-based therapy in neurology. Tissue Eng Part A. 2014;20:1794–806.
Bharadwaj S, Liu G, Shi Y, Wu R, Yang B, He T, et al. Multipotential differentiation of human urine-derived stem cells: potential for therapeutic applications in urology. Stem Cells. 2013;31:1840–56.
Xin H, Li Y, Chopp M. Exosomes/miRNAs as mediating cell-based therapy of stroke. Front Cell Neurosci. 2014;8:377.
De Jong OG, Van Balkom BW, Schiffelers RM, Bouten CV, Verhaar MC. Extracellular vesicles: potential roles in regenerative medicine. Front Immunol. 2014;5:608.
Fu Y, Guan J, Guo S, Guo F, Niu X, Liu Q, et al. Human urine-derived stem cells in combination with polycaprolactone/gelatin nanofibrous membranes enhance wound healing by promoting angiogenesis. J Transl Med. 2014;12:274.
Hu GW, Li Q, Niu X, Hu B, Liu J, Zhou SM, et al. Exosomes secreted by human-induced pluripotent stem cell-derived mesenchymal stem cells attenuate limb ischemia by promoting angiogenesis in mice. Stem Cell Res Ther. 2015;6:10.
Jiang ZZ, Liu YM, Niu X, Yin JY, Hu B, Guo SC, et al. Exosomes secreted by human urine-derived stem cells could prevent kidney complications from type I diabetes in rats. Stem Cell Res Ther. 2016;7:24.
Barquilla A, Pasquale EB. Eph receptors and ephrins: therapeutic opportunities. Annu Rev Pharmacol Toxicol. 2015;55:465–87.
Goldshmit Y, Galea MP, Bartlett PF, Turnley AM. EphA4 regulates central nervous system vascular formation. J Comp Neurol. 2006;497:864–75.
Ling KK, Jackson M, Alkam D, Liu D, Allaire N, Sun C, et al. Antisense-mediated reduction of EphA4 in the adult CNS does not improve the function of mice with amyotrophic lateral sclerosis. Neurobiol Dis. 2018;114:174–83.
Wei HX, Yao PS, Chen PP, Guan JH, Zhuang JH, Zhu JB, et al. Neuronal EphA4 regulates OGD/R-induced apoptosis by promoting alternative activation of microglia. Inflammation. 2019;42:572–85.
Fukuhara S, Sako K, Noda K, Nagao K, Miura K, Mochizuki N. Tie2 is tied at the cell-cell contacts and to extracellular matrix by angiopoietin-1. Exp Mol Med. 2009;41:133–9.
Deshors P, Toulas C, Arnauduc F, Malric L, Siegfried A, Nicaise Y, et al. Ionizing radiation induces endothelial transdifferentiation of glioblastoma stem-like cells through the Tie2 signaling pathway. Cell Death Dis. 2019;10:816.
Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78:838–48.
Ge XT, Lei P, Wang HC, Zhang AL, Han ZL, Chen X, et al. miR-21 improves the neurological outcome after traumatic brain injury in rats. Sci Rep. 2014;4:6718.
Cai, Wei D, Chen S, Chen X, Li S, Chen W, et al. MiR-145 protected the cell viability of human cerebral cortical neurons after oxygen-glucose deprivation by downregulating EPHA4. Life Sci. 2019;231:116517.
Acknowledgements
The authors would like to acknowledge the helpful comments on this paper received from the reviewers.
Author information
Authors and Affiliations
Contributions
WP designed the study. WP, XHX, and MZ collated the data, carried out data analyses, and produced the initial draft of the manuscript. XYS contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pan, W., Xu, X., Zhang, M. et al. Human urine-derived stem cell-derived exosomal miR-21-5p promotes neurogenesis to attenuate Rett syndrome via the EPha4/TEK axis. Lab Invest 101, 824–836 (2021). https://doi.org/10.1038/s41374-021-00574-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00574-w
This article is cited by
-
Urine-derived mesenchymal stem cells-derived exosomes enhances survival and proliferation of aging retinal ganglion cells
BMC Molecular and Cell Biology (2023)
-
Bioengineering extracellular vesicles as novel nanocarriers towards brain disorders
Nano Research (2023)
-
Human Oral Mucosa Stem Cells Increase Survival of Neurons Affected by In Vitro Anoxia and Improve Recovery of Mice Affected by Stroke Through Time-limited Secretion of miR-514A-3p
Cellular and Molecular Neurobiology (2023)