Abstract
The tumor microenvironment is increasingly recognized as key player in cancer progression. Investigating heterotypic interactions between cancer cells and their microenvironment is important for understanding how specific cell types support cancer. Forming the vasculature, endothelial cells (ECs) are a prominent cell type in the microenvironment of both normal and neoplastic breast gland. Here, we sought out to analyze epithelial–endothelial cross talk in the breast using isogenic non-tumorigenic vs. tumorigenic breast epithelial cell lines and primary ECs. The cellular model used here consists of D492, a breast epithelial cell line with stem cell properties, and two isogenic D492-derived EMT cell lines, D492M and D492HER2. D492M was generated by endothelial-induced EMT and is non-tumorigenic while D492HER2 is tumorigenic, expressing the ErbB2/HER2 oncogene. To investigate cellular cross talk, we used both conditioned medium (CM) and 2D/3D co-culture systems. Secretome analysis of D492 cell lines was performed using mass spectrometry and candidate knockdown (KD), and overexpression (OE) was done using siRNA and CRISPRi/CRISPRa technology. D492HER2 directly enhances endothelial network formation and activates a molecular axis in ECs promoting D492HER2 migration and invasion, suggesting an endothelial feedback response. Secretome analysis identified extracellular matrix protein 1 (ECM1) as potential angiogenic inducer in D492HER2. Confirming its involvement, KD of ECM1 reduced the ability of D492HER2-CM to increase endothelial network formation and induce the endothelial feedback, while recombinant ECM1 (rECM1) increased both. Interestingly, NOTCH1 and NOTCH3 expression was upregulated in ECs upon treatment with D492HER2-CM or rECM1 but not by CM from D492HER2 with ECM1 KD. Blocking endothelial NOTCH signaling inhibited the increase in network formation and the ability of ECs to promote D492HER2 migration and invasion. In summary, our data demonstrate that cancer-secreted ECM1 induces a NOTCH-mediated endothelial feedback promoting cancer progression by enhancing migration and invasion. Targeting this interaction may provide a novel possibility to improve cancer treatment.
Similar content being viewed by others
Introduction
Organ morphogenesis is dependent on heterotypic interactions between multiple cell types. In breast morphogenesis, the epithelial compartment generates branching ducts that result in terminal duct lobular units (TDLU) [1]. Ducts and TDLUs are surrounded by a basement membrane, embedded in stroma consisting of extracellular matrix (ECM) as well as multiple cell types including fibroblasts, immune cells, and endothelial cells (ECs) forming the microvessels [1, 2]. Formation of TDLU is highly dependent on heterotypic interactions between the epithelial cells and the surrounding vascular-rich stroma. Multiple studies have shown that the stromal compartment plays a fundamental role when it comes to epithelial morphogenesis [2,3,4,5,6]. For example, the ability of breast epithelial cells to form TDLU-like structures in a 3-dimensional environment in vitro is strongly enhanced by the presence of breast ECs [2]. In addition to its role in normal development, recent studies provide evidence that proliferation and migration of cancer cells is also largely dependent on interactions with the surrounding stroma [3, 7,8,9].
The tumor microenvironment is composed of a plethora of different cell types and ECM [10]. The interaction between cancer cells and cells of the microenvironment is a crucial determinant of cancer progression. Co-evolution of cancer cells and tumor stroma can lead to the generation of niches that support tumor growth, both at the primary site and at distant metastatic sites [11]. In this context, many studies have been focusing on identifying the role of tumor-associated macrophages and cancer-associated fibroblasts in cancer progression [12,13,14,15,16].
Recent research, however, suggests that ECs of blood vessels play a role in cancer progression that is far beyond delivering oxygen and nutrients [7, 17,18,19,20]. There is evidence that the endothelium can impact cancer progression by either preventing or supporting tumor growth and metastasis formation. An intact vasculature can keep metastatic cells in a dormant and non-proliferative state whereas a sprouting vasculature rather supports metastatic outgrowth [7, 21, 22]. Ghajar and colleagues found that endothelial-derived thrombospondin-1 induces cancer cell dormancy in breast cancer and therefore prevents outgrowth of metastasis in lung and bones [7]. However, in the presence of sprouting neovasculature this suppressive effect was lost and the endothelium now appeared to promote tumor outgrowth via TGF-b1 and periostin [7]. Therefore, an essential question to ask is how cancer cells interact with ECs in order to promote the generation of a vascular niche promoting tumor growth and cancer progression. Presently, a number of molecular mediators of angiogenesis have been identified [23]. The most commonly described candidates include vascular endothelial growth factor (VEGF), fibroblast growth factor 2 (FGF2), tumor necrosis factor alpha (TNF-a), transforming growth factor beta 1 (TGF-b1), platelet-derived growth factor (PDGF), as well as angiopoietins. Laughner et al. were able to show that HER2 signaling in breast cancer cells could increase HIF1a-mediated VEGF expression resulting in increased tumor angiogenesis [24]. Lee et al. showed that increased IL-6 secretion in breast cancer cells induces secretion of CCL5 in lymphatic endothelial cells (LEC), which in return enhances cancer cell migration [18]. Furthermore, cancer cell-secreted IL-6 increases VEGF expression in LECs, which enhances lung vascular permeability and lymph node angiogenesis and thereby promotes metastatic outgrowth [18, 19]. In addition, the NOTCH signaling pathway has recently been linked to angiogenesis [23, 25, 26]. Murtas et al. report increased NOTCH1 expression by tumor endothelium in cutaneous melanoma that was linked to increasing microvascular density [25]. This suggests that targeting the interaction between cancer cells and their niches may provide additional means to inhibit progression of cancer. However, our understanding of the generation of vascular niches and molecular interactions within the niches is generally still rudimentary.
We have previously established a breast epithelial cell line with stem cell properties, referred to as D492 [27,28,29,30]. D492 can generate both luminal and myoepithelial cells, and in 3D rBM (reconstituted basement membrane, matrigel) culture it forms branching structures reminiscent of TDLU in vivo. When D492 was co-cultured with ECs in an organoid culture, a subpopulation of D492 underwent EMT and such a structure isolated gave rise to D492M [27]. D492HER2 was generated by overexpressing the ErbB2/HER2 oncogene in D492, which then underwent oncogene-induced EMT [31]. In contrast to D492M, D492HER2 has tumorigenic properties, as evidenced by injection of cells in the mammary fat pads of NOD scid gamma (NSG) mice. The different tumorigenicities of these two isogenic EMT derivatives of D492 present an interesting research platform to study tumorigenicity, especially with regard to interaction with the endothelium.
In this study, we show that conditioned medium (CM) from D492HER2 increases endothelial network formation in vitro and induces an endothelial feedback promoting D492HER2 migration and invasion. Secretome analysis identified extracellular matrix protein 1 (ECM1) as potential pro-angiogenic factor in D492HER2. Recombinant ECM1 (rECM1) increases endothelial network formation while knockdown (KD) of ECM1 reduces the ability of D492HER2-CM to increase the endothelial network. ECM1 has previously been associated with decreased overall and distant metastasis-free survival when expressed in HER2+ breast tumors [32,33,34]. A role in angiogenesis and cancer cell migration and invasion has been suggested in laryngeal carcinoma, cholangiocarcinoma, gastric cancer, bladder cancer, and breast cancer [32, 35,36,37,38]. However, the mechanism through which ECM1 induces a pro-tumorigenic vascular niche promoting cancer progression is still unknown. Finally, increased expression of NOTCH1 and NOTCH3 in ECs treated with D492HER2-CM or rECM1 indicates a possible novel modulatory role of ECM1 in NOTCH signaling during angiogenesis and cancer.
Materials and methods
Cell culture
D492, D492M, and D492HER2 cells were maintained in H14 medium in tissue culture-treated T25 Falcon flasks (BD Biosciences (BD), Franklin Lakes, NJ, USA) coated with collagen I (Advanced Biomatrix, San Diego, CA, USA), as described previously [27, 31]. The cells were grown at 37 °C and 5% CO2 and subcultured into new flasks 1–2 times per week in a ratio of 1:10 (D492), 1:5 (D492M), or 1:15 (D492HER2). MDA-MB-231 was cultured in Gibco™ RPMI 1640 medium containing 10% fetal bovine serum (FBS) (Invitrogen), further referred to as R10F, and MCF-7 in Gibco™ DMEM:F12 medium with 10% FBS. Both cell lines were subcultured 1–2 times per week at a ratio of 1:10. Primary human umbilical vascular ECs (HUVEC) were obtained from the National University Hospital (Landspitali) Reykjavik and cultured up to passage 8 in EBM2 medium (Lonza) supplemented with growth factors and 5% FBS, further referred to as EGM5. HUVECs were subcultured 1–2 times per week at a ratio of 1:10. Culture medium was changed three times per week and routinely checked for mycoplasma contamination.
CM and mass spectrometry
D492, D492M, and D492HER2 were grown in H14 medium until 70–80% confluence. Cells were washed with 1x PBS, and half of the original volume of fresh H14 was added in order to enrich for secreted factors. After 48 h incubation at 37 °C, CM was collected, centrifuged at 2000 rpm for 3 min and sterile filtered through 0.22 µm filter unit. For endothelial network formation in 3D culture, 10,000 HUVECs were seeded in freshly collected CM (or unconditioned control H14) and fresh EGM5 medium (ratio 1:1) in a 96 well angiogenesis plate (#89646, Ibidi) on top of 10 µl solidified rBM Matrigel (#354230, Corning). For recombinant protein experiments, rECM1 (#TP723147, Origene) was added to the medium in a final concentration of 15, 30, 60, or 120 ng/ml. 60 ng/ml was then chosen as concentration for further experiments. For endothelial NOTCH inhibition, gamma-secretase inhibitor tert-Butyl (2S)-2-[[(2S)-2-[[2-(3,5-difluorophenyl)acetyl]amino]propanoyl]amino]-2-phenylacetate (DAPT) (#D5942, Sigma-Aldrich) was added to the medium in a final concentration of 20 µM. Phase-contrast images of the establishing endothelial network were taken after 4, 24, and 48 h. Images were converted to RGB color and analyzed using ImageJ Angiogenesis Analyzer macro. For mass spectrometry, D492 lines were grown in T175 flasks and CM was collected as described above, concentrated for 55 min using EMD Millipore Amicon™ Ultra-15 Centrifugal Filter Units (#UFC900324, Merck Millipore) followed by buffer exchange to 100 mM TRIS/HCL buffer. Samples (triplicates) were stored at −80 °C. Label-free relative protein quantification (LFQ) by nLC MS/MS after trypsin digestion was performed at the FingerPrints Proteomics Facility, University of Dundee, UK, and raw data were analyzed using MaxQuant software (version 1.6.2.1). Quantitative and statistical analysis was performed using XLStat (version 2018.1). Data were p value corrected (significance level 0.05) and sorted based on greter than or equal to twofold higher secretion (LFQ intensity) by D492HER2 compared with both D492 and D492M. The resulting candidate list of 77 proteins was used for GO term analysis using PANTHER database (statistical overrepresentation test). As annotation dataset we used “GO biological process complete” with Bonferroni correction for multiple testing. To investigate feedback effects of conditioned/induced ECs, CM from D492 lines was added to 40–50% confluent HUVECs (as described above) and incubated for 48 h. Cells were washed with 1x PBS, and fresh EGM5 (half volume) was added to collect conditioned endothelial-secreted factors. After 48 h, CM from induced vs. non-induced HUVECs was collected, centrifuged (2000 rpm, 3 min) and filtered (0.22 µm) and added to 40% confluent D492HER2 together with fresh H14 (ratio 1:1) for 48 h.
Migration and invasion assay
Migration was analyzed using transwell-filter units for 24-well plates with 8 µm pore size (#353097, Corning). Thirty thousand cells were seeded on top of the filter in 250 µl culture medium. For the invasion assay, filters were pre-coated with 1:10 rBM in H14 for 30 min at 37 °C. For the migration assay, cells were incubated for 24 h and for 48 h in the invasion assay. rECM1 and NOTCH inhibitor DAPT were used in the concentrations as described before. For migration towards ECs, HUVECs were grown in 500 µl EGM5 to 80% confluence in 24-well plate. Then, medium was changed to H14+ EGM5 (1:1), and D492 line cells were seeded on top of filter units (triplicates) in H14+ EGM5 (1:1). As positive control, 10% FBS was added to the medium below the filter instead of HUVECs and as negative control, medium above and below the filter unit was identical (H14+ EGM5, 1:1). For migration and invasion of D492HER2 treated with CM of induced vs. non-induced ECs, 30,000 D492HER2 cells were seeded in 250 µl H14 on top of the non-coated or rBM-coated filter units (triplicates) and 500 µl H14 containing 10% FBS as attractant was added below. For MDA-MB-231, R10F was used instead of H14 while keeping the same experimental setup. After 24 h or 48 h, non-migratory or non-invasive cells were removed from the top of the filter using Q-tips, migratory cells below the filter were fixed for 10 min with 4% PFA (#252549, Sigma-Aldrich) and stained for 30 min with 1:5000 diluted DAPI nuclear staining (20 mg/ml) (#D9542, Sigma-Aldrich). Using an EVOS FL Auto 2 Cell Imaging System (Thermo Fisher Scientific), three images per replicate were taken at ×10 magnification. Images were converted to 8-bit in ImageJ (version 2.0.0), threshold-adjusted and binary-converted. Migratory cells were counted using the “analyze particles” function.
Transient KD using siRNA
KD was performed in 24-well plate using pre-designed Silencer® Select human ECM1 siRNA (#s4441, Thermo Fisher Scientific) compared with a scrambled negative control (Silencer Select negative control No. 1, Thermo Fisher Scientific, #4390843). Cells were reverse transfected for 48 h according to the manufacturer’s protocol using a siRNA concentration of 10 nM. Proliferation was monitored using an IncuCyte ZOOM 2016B System. KD was confirmed using RT-qPCR and western blotting. After 48 h, medium was changed to normal H14 and incubated for another 48 h for collection of CM as described above. Endothelial network formation was analyzed as previously described for HUVECs treated with CM from D492HER2siECM1 compared with control D492HER2.
Generation of stable KD and overexpression cell lines using CRISPRi/CRISPRa
In order to generate stable ECM1 KD in D492HER2 and overexpression in D492 and D492M, we used the novel CRISPRi (inhibition) and CRISPRa (activation) system. sgRNA plasmids were purchased from Genscript; for ECM1 overexpression and KD, a combination of two sgRNAs was used. Overexpression sgRNAs were pre-design and located between 1 and 200 bp upstream of the ECM1 transcription start site and KD sgRNAs custom-designed to bind between 1 and 200 bp downstream. sgRNAs were cloned into the plenti sgRNA(MS2)_zeo plasmid (Genscript). As negative control, an empty plenti sgRNA(MS2)_zeo plasmid was used (Genscript). For production of lentiviral particles, HEK293T cells were used and virus was collected 48 and 72 h after transfection. D492 and D492M were first transduced with plenti dCAS9-VP64-Blast plasmid for transcriptional activation and selected with blasticidin at a concentration of 2 µg/ml for 7 days. D492HER2 were transduced with pHR-SFFV-KRAB-dCas9-P2A-mCherry plasmid for repression and sorted for mCherry expression. Next, transduction with lentivirus containing sgRNA plasmids for overexpression (D492 and D492M containing dCas9-VP64 cassette) and KD (D492HER2 containing dCas9-KRAB cassette) was performed. For both KD and OE, two different gRNAs were pooled (KD: ECM1 custom gRNA 1 and 2 (GTGGTCAGTTGCCCCAGGAT, GCCGGCCACTGAAGCTTGTC) and OE: ECM1 SAM guide RNA 1 and 2 (CATCTACAGGCTGCCTTCTG, GAAACTGAGGCACAAACTAG)). Cells were selected with 400 µg/ml (D492, D492M) or 600 µg/ml (D492HER2) zeocin (Invitrogen) for 14 days. For enhancement of ECM1 OE, D492 and D492M were then transduced with lenti MS2-P65-HSF1 Hygro plasmid, containing enhancer domains MS2, P65, and HSP1 that bind to VP64 and enhance its activation activity. Cells were selected with 100 µg/ml hygromycin (Sellekchem, #S2908) for 10–12 days. ECM1 KD or OE were validated on gene expression level using RT-qPCR.
RNA microarray
For RNA microarray analysis, D492, D492M, and D492HER2 with and without ECM1 overexpression/KD were grown until 80% confluence, fresh H14 medium was added and CM was collected after 48 h. HUVECs were grown to 40% confluence, washed with 1x PBS and CM was added together with fresh EGM5 (ratio 1:1). After 24 h, RNA was extracted using the RNeasy Mini Kit (Qiagen). Sample concentration and quality was analyzed using the Agilent 2100 Bioanalyzer System (Agilent). As RNA microarray, Affimetrix Human Clariom S Assay was performed at the sequencing core facility, German Cancer Research Center (DKFZ) Heidelberg, Germany. Raw data were analyzed using Chipster high-throughput data analysis software v3.12.
RNA, cDNA, and RT-qPCR
Total RNA was extracted with Trizol (Life Technologies) and reverse transcribed with hexanucleotides using the SuperScript® IV First-Strand Synthesis System (Invitrogen). Resulting cDNA (10 ng per reaction) was used for quantitative real-time PCR, in master mix (Life Technologies) with pre-designed primer pairs ECM1 (Hs.PT.58.20438560), NOTCH1 (Hs.PT.58.23074795), NOTCH3 (Hs.PT.58.38492200), and beta-2-microglobulin (B2M) as reference gene (Hs.PT.58v.18759587). Experiments were carried out in triplicate on the 7500 Real-Time PCR System (Life Technologies). Expression levels were normalized to the reference gene, and relative mRNA differences were calculated with the ΔΔCt method according to the “Minimum Information for Publication of Quantitative Real-Time PCR Experiments” (MIQE) guidelines [39].
Protein isolation and western blotting
Protein was isolated using RIPA lysis buffer supplemented with phosphatase and protease inhibitor cocktails (Life Technologies). For western blotting, 5–10 μg protein was used per lane, unless otherwise stated. Samples were denatured using 10% mercaptoethanol at 95 °C for 5 min and run on NuPage 10% Bis-Tris gels (Life Technologies) in 2-(N-morpholino)ethanesulfonic acid running buffer. Samples were then transferred to immobilon FL polyvinylidenfluorid (PVDF) membranes (Millipore). Membranes were blocked in Li-Cor blocking buffer, and primary antibodies (ECM1: sc-515843 (Santa Cruz), beta tubulin: ab6046 (Abcam)) were incubated overnight at 4 °C. Near-infrared fluorescence visualization was measured using Odyssey CLx scanner (Li-Cor, Cambridge, UK).
In vivo tumor formation assay
To assess tumorigenicity of D492HER2 and D492HER2 with KD of ECM1, cells were injected subcutaneously into female NSG mice, bred at the Department of Comparative Medicine, Oslo University Hospital. The mice were kept in pathogen-free environment at a constant temperature (21.5 ± 0.5 °C) and humidity (55 ± 5%); 15 air changes/h and a 12 h light/dark cycle. The animals were 5–6 weeks old and their weight was 18–20 g before they were included in experiments. Anesthesia was obtained with 5% (v/v) Sevofluran along with 1 L oxygen and 3 L nitrous oxide, given with inhalation mask. Food and water were supplied ad libitum. 5 × 105 cells in 100 µl of 1:1 mixture of PBS and Matrigel (Corning, #354248) were injected on both flanks of the animal. Tumor growth was measured twice a week using a caliper, and the tumor volume was calculated according to the formula 0.5 × length × width2. Animals were sacrificed performing cervical dislocation at the end of the experiments, or when tumor volume reached 1500 mm3, the weight loss exceeded 20% or when they became moribund. Tumors were dissected and from each tumor one part was frozen in liquid nitrogen and the other was fixed in PFA and embedded in paraffin. All experiments involving animals have been approved by the Norwegian Animal Research Authority (ethical approval FOTS ID: 12080) and conducted according to the regulations of the Federation of European Laboratory Animals Science Association (FELASA) [40].
Immunofluorescence (IF) staining
HUVECs were grown to 80% confluence on eight-well chamber slides (#354108, Falcon), fixed with 3.7% PFA, permeabilized with 0.1% Triton X-100, and unspecific background was blocked using 10% FBS in 1x PBS. Incubation with primary antibodies for ECM1 (Sigma-Aldrich, #HPA027241), NOTCH1 (#sc-376403, Santa Cruz), and NOTCH3 (#ab23426, Abcam) was done overnight at 4 °C, followed by incubation with secondary antibodies conjugated with Alexa Fluor-488, -546, or -647 for 1 h at RT. For nuclei staining DAPI was used together with secondary antibodies at a concentration of 1:5000. Imaging was done using FV1200 Olympus inverted confocal microscope. For IHC staining, paraffin-embedded tumor tissue samples were processed in xylene and ethanol in order to remove paraffin and then rehydrated in dH2O. Following rehydration, high-temperature antigen retrieval in 1x T/E buffer was performed and samples were stained as described above. Counterstaining was performed with filtered hematoxylin for 1–2 min.
Statistical analysis
All experiments were performed at minimum in triplicate. Graphs were generated in Microsoft Excel 2015. Error bars represent the standard deviation of the mean. Statistical analysis was performed in R 3.3.3. (R Development Core team, 2013). Data were checked for normal distribution and Studentʼs t test or ANOVA followed by Tukey’s honest significant difference test on linear models was performed when normally distributed whereas Kruskal–Wallis chi-squared test was used when not normally distributed. The significance level was 0.05.
Results
CM from D492HER2 promotes endothelial network formation
Initially, we aimed to analyze the heterotypic interactions between normal and cancerous epithelial cells of the mammary gland and ECs. To address this, we used isogenic normal and cancerous cell lines derived from the D492 breast epithelial progenitor cell line [28, 30]. We collected CM from D492, D492M, and D492HER2 and analyzed the effect on endothelial network formation in vitro. ECs were seeded in CM from D492, D492M, and D492HER2 on top of rBM, and endothelial network formation was analyzed after 4, 24, and 48 h (Fig. 1a, b, Supplementary Fig. S1). Assessment of EC viability confirmed similar cell viability upon CM treatment compared with H14 ctrl medium (Supplementary Fig. S13). The established network of ECs treated with CM from D492HER2 but not from D492 and D492M showed significantly higher numbers of master junctions, master segments, meshes, and higher total master segment length after 4 h (Fig. 1a, b). This indicates that D492HER2-CM enhances early endothelial network formation in vitro. All quantified parameters were also significantly higher upon treatment with D492HER2-derived CM after 24 and 48 h indicating stability and persistence of the observed pro-angiogenic effect of D492HER2-CM over time (Supplementary Fig. S1).
a Enhanced endothelial network formation in D492HER2-CM after 4 h. Top: representative phase-contrast images of HUVEC endothelial network after 4 h on top of rBM in control (unconditioned) medium and conditioned medium from D492, D492M, and D492HER2. Each condition included 4–6 wells as replicates and 1–2 images were taken per well at ×10 magnification (scale bar = 100 µm). Bottom: corresponding angiogenesis images (ImageJ, angiogenesis analyzer plugin) showing branches, junctions, master junctions, master segments, and meshes. b Endothelial network formation—quantification. Quantification of master junctions, master segments total master segment length and meshes for each image using angiogenesis analyzer plugin (average and standard deviation per condition). Statistical analysis performed in R, one-way ANOVA, ***p < 0.001, **p < 0.01, *p < 0.05, <0.1. c Endothelial feedback—schematic overview. Schematic workflow of conditioning of ECs (see “Materials and methods”). d D492HER2-induced ECs promote D492HER2 migration and invasion. Transwell-migration and invasion assays of D492HER2 treated with unconditioned medium, unconditioned HUVEC-CM, D492-conditioned HUVEC-CM, and D492HER2-conditioned HUVEC-CM. Number of migratory/invasive cells is shown for the different treatments (Studentʼs t test, ***p < 0.001, **p < 0.01, *p < 0.05, <0.1). e Migration toward ECs—schematic overview. Schematic setup of the transwell-migration assay of D492, D492M, and D492HER2 toward HUVECs seeded below the transwell filter. Migration toward HUVECs (in H14 + EGM1) was compared with migration toward 10% FBS in H14 + EGM5 (pos. ctrl) and plain H14 + EGM5 (neg. ctrl). f Increased D492HER2 migration toward ECs. Migration of D492, D492M, and D492HER2 toward HUVECs. Number of migratory cells is shown for the different treatments (Studentʼs t test, ***p < 0.001, **p < 0.01, *p < 0.05, <0.1).
D492HER2-induced ECs feedback on D492HER2 by promoting migration and invasion
After identifying D492HER2 as a cell line enhancing endothelial network formation, we collected CM from D492HER2-induced ECs and treated D492HER2 (Fig. 1c) in order to investigate a possible feedback response through cellular cross talk. Interestingly, we observed that treatment with CM from D492HER2-induced ECs stimulated increased migration and invasion of D492HER2, in contrast to CM from untreated ECs and D492-induced ECs (Fig. 1d), suggesting a positive feedback. To identify whether migration of D492HER2 cells was specifically directed toward the endothelium rather than being general, we performed indirect co-culture of D492, D492M, and D492HER2 in a transwell assay with or without ECs present below the filter (Fig. 1e). When analyzing the number of migratory cells, D492HER2 in particular showed an increased migratory phenotype toward ECs below the filter (Fig. 1f), supporting the hypothesis that migration is increased toward the endothelium mediated by induced ECs. In line with this, we could see significantly more D492HER2 than D492 or D492M cells directly associated with ECs in 2D and 3D co-culture with ECs (Supplementary Fig. S2).
Enrichment of angiogenesis-regulating proteins and identification of ECM1 in D492HER2 secretome
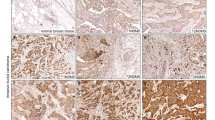
In order to identify secreted factors from D492HER2 that are mediating the observed pro-angiogenic effect, we performed label-free mass spectrometry on concentrated CM from all three D492 cell lines. In order to determine candidates involved in tumorigenicity rather than EMT phenotype, we focused on proteins higher secreted by D492HER2 compared with both D492 and D492M. We used RT-qPCR and transcriptomic data to verify secretome candidates. After correction of the data for significance (0.05 significance level) and at least twofold difference in secretion, we received a list of a total of 77 proteins more highly secreted by D492HER2 compared with D492 and D492M (Fig. 2a and Supplementary Table S1). GO term analysis showed 6.7-fold enrichment for GO term group “regulation of angiogenesis” (GO:0045765) among D492HER2-secreted candidates (Fig. 2b), consisting of seven gene members (Fig. 2c). We then compared secretion levels of these seven candidates in our secretome dataset and identified ECM1 and chitinase 3-like-1 (CHI3L1) as the two proteins with the most prominent secretion profile (Fig. 2d). Interestingly, secretion levels of commonly recognized angiogenesis inducers such as VEGFa, FGF2, or PDGF were not significantly elevated in our cell lines, suggesting a different mechanism behind the observed pro-angiogenic effect of D492HER2 secretome (Fig. 2d). CHI3L1, also known as YKL-40, is a secreted glycoprotein, which is associated with a number of biological functions and also plays a role in chronic inflammation diseases and cancer [41,42,43]. Recently, it has been linked to idiopathic pulmonary fibrosis [44]. Previous studies provide evidence for a role of YKL-40 in tumor angiogenesis by mediating VEGF signaling [45, 46]. ECM1 is a secreted glycoprotein, which was first identified in the mouse osteogenic stromal cell line MN7 [47]. It was initially described as modulator of proliferation and differentiation of epidermal keratinocytes and basement membrane reconstitution in the skin [48]. However, it has also been linked to angiogenesis and malignant transformation [32,33,34, 37, 48,49,50]. Interestingly, analysis to determine correlation to different gene modules using GOBO gene set analysis database (University of Lund, Sweden) revealed a significant Spearman correlation of ECM1 to the gene module “stroma” (Fig. 2e). Kaplan–Maier survival analysis revealed for both CHI3L1 and ECM1, a significant correlation of gene expression with decreased survival (Supplementary Fig. S4). However, whereas in case of CHI3L1 this correlation is not specifically associated with a certain cancer subtype, ECM1 expression is significantly correlated with decreased distant metastasis-free survival (DMSF) when expressed in HER2+ and estrogen receptor-negative (ER−) breast tumors (Fig. 2f, g, Supplementary Figs. S3 and S4) [49]. These data put together led our focus on ECM1 as the main candidate for further investigation in the context of stromal interaction.
a 77 candidates. Heat map of 77 proteins higher secreted by D492HER2 compared with both D492 and D492M (label-free mass spectrometry secretome data, p < 0.05, threshold fold change greater than or equal to twofold). b Enrichment of angiogenesis-regulating genes in D492HER2 secretome (GO terms). Enrichment of “Regulation of angiogenesis” GO term group GO:0045765 among D492HER2 secretome. Statistical overrepresentation test (Panther DB) was performed using “GO biological process complete” as annotation dataset with Bonferroni correction for multiple testing. c GO:0045765 gene members. Proteins part of GO term group GO:0045765 (Regulation of angiogenesis). d Highest secretion levels of ECM1 and CHI3L1. Secretion levels (label-free quantification LFQ) of the seven candidates from c (left of dotted line) compared WITH secretion levels of commonly known angiogenesis inducers (right of dotted line). e Correlation of ECM1 to eight gene modules. Spearman correlation of ECM1 expression to different gene modules in breast tumor samples (GOBO Genset analysis, Lund University). f DMFS in ECM1+/HER2+ patients. Kaplan–Meier plot of ECM1 in HER2+ tumors showing correlation of high expression with decreased distant metastasis-free survival (DMFS). g ECM1 expression in HER2+ tumors. ECM1 expression in representative HER2+ tumor samples showing low and high expression. Cells were counterstained with hematoxilyn. Scale bar = 100 µm.
ECM1 enhances endothelial network and induces endothelial feedback
To verify that ECM1 is involved in the inducing effects of D492HER2 on endothelial network formation and positive feedback, we investigated whether rECM1 is capable of inducing network formation and feedback as well. We treated ECs with 15, 30, 60, and 120 ng/ml of rECM1 and quantified network formation as described previously. Indeed, rECM1 had significantly increased the endothelial network formed after 4 and 24 h as quantified by numbers of master junctions, meshes, master segments, and total master segment length (Fig. 3a, b). Hereby, the strongest effect was seen for treatment with 30 and 60 ng/ml rECM1 (Fig. 3b). This indicates that the system may already be saturated at these concentrations rather than rECM1 having a dose-dependent effect. To look at the endothelial feedback, we induced ECs with 60 ng/ml rECM1 and treated D492HER2 with CM of ECM1-induced ECs (Fig. 3c). CM of ECM1-induced ECs significantly increased D492HER2 migration and invasion (Fig. 3d), confirming that ECM1 does play a role in inducing the endothelial feedback induced by D492HER2.
a Increased endothelial tube formation with recombinant ECM1. Top: HUVEC endothelial network on top of matrigel in different concentrations of recombinant ECM1 (after 24 h). 4–6 wells as replicates per condition and 1–2 images taken per well at ×10 magnification (scale bar = 100 µm). Bottom: corresponding angiogenesis images (ImageJ angiogenesis analyzer). b Endothelial network formation—quantification. Quantification of master junctions, master segments, total master segment length, and meshes for each image using angiogenesis analyzer plugin. One-way ANOVA, ***p < 0.001, **p < 0.01, *p < 0.05, <0.1. c Endothelial feedback by rECM1—schematic overview. Schematic workflow of conditioning of ECs using 60 ng/ml rECM1. d rECM1-induced ECs promote D492HER2 migration and invasion. Transwell-migration and invasion assays of D492HER2 treated with unconditioned medium, unconditioned HUVEC-CM and 60 ng rECM1-treated HUVEC-CM. Number of migratory/invasive cells shown for different treatments (Studentʼs t test, ***p < 0.001, **p < 0.01, *p < 0.05, <0.1).
To verify the activity of ECM1, we induced stable overexpression of ECM1 in D492 and D492M. CM from D492 and D492M stably overexpressing ECM1 showed pro-angiogenic properties (Fig. 4a, b). Furthermore, D492 and D492M overexpressing ECM1 show increased migration toward ECs, similar as observed for D492HER2 (Supplementary Fig. S6). Next, we generated transient KD of ECM1 in D492HER2 using siRNA (Supplementary Fig. S5) and stable KD using CRISPRi. Both transient and stable KD were successful (over 90% KD), as verified on gene as well as protein level (Supplementary Figs. S5 and S6). In accordance with the observed effects of rECM1 and ECM1 overexpression, treatment with CM from D492HER2-kdECM1 resulted in significantly lower endothelial network formation compared with control D492HER2 (Fig. 4a, b). Similar results were obtained with CM from D492HER2 with transient ECM1 KD (Supplementary Fig. S5). In addition to these results on the D492 cell lines, we investigated pro-angiogenic properties of SKBR3 (HER2+ and ECM1−) and HCC202 (HER2+ and ECM1+) breast cancer cell lines. Network formation data showed increased pro-angiogenic properties of HCC202-CM but not SKBR3-CM compared with controls, confirming the specific role of ECM1 in this context (Supplementary Fig. S10). Analysis of the endothelial feedback showed that treatment of ECs with CM from D492HER2-kdECM1 did not induce an endothelial response to D492HER2 increasing cell migration and invasion (Fig. 4c, d). Also, the observed increased migration of D492HER2 toward ECs (Fig. 4c) could be depleted upon KD of ECM1, supporting the role of ECM1 in mediating the endothelial feedback and promoting migration toward the endothelium (Fig. 4e, f). These data suggest ECM1 as a specific mediator of endothelial response promoting cancer progression.
a Reduced pro-angiogenic properties of D492HER2-CM upon ECM1 knockdown. Top: HUVEC endothelial network on top of matrigel in CM from D492 and D492M with ECM1 overexpression and D492HER2 with ECM1 knockdown compared with empty vector control (after 4 h). 4–6 wells as replicates per condition and 1–2 images taken per well at ×10 magnification (scale bar = 100 µm). Bottom: corresponding angiogenesis images (ImageJ angiogenesis analyzer). b Endothelial network formation—quantification. Quantification of master junctions, master segments, total master segment length, and meshes for each image using angiogenesis analyzer plugin. One-way ANOVA, ***p < 0.001, **p < 0.01, *p < 0.05, <0.1. c Endothelial feedback reduction—schematic overview. Schematic workflow of conditioning of ECs using CM from D492HER2-kdECM1. d No endothelial feedback on D492HER2 migration upon ECM1 knockdown. Transwell-migration and invasion assays of D492HER2 treated with unconditioned medium, unconditioned HUVEC-CM, D492HER2-conditioned HUVEC-CM, and D492HER2-kdECM1-conditioned HUVEC-CM. Studentʼs t test, ***p < 0.001, **p < 0.01, *p < 0.05, <0.1. e Migration toward ECs—schematic overview. Schematic setup of the transwell-migration assay of D492HER2 empty ctrl. and D492HER2-kdECM1 toward HUVECs compared with pos. ctrl (10% FBS) and neg. ctrl (plain H14 + EGM5). f No increased migration toward ECs upon ECM1 knockdown. Migration of D492HER2 empty ctrl and D492HER2-kdECM1 toward HUVECs. Number of migratory cells for different treatments (Studentʼs t test, ***p < 0.001, **p < 0.01, *p < 0.05, <0.1).
Subcutaneous injection of D492HER2 ctrl and D492HER2-kdECM1 into mice indicated a reduction in tumor volume upon ECM1 KD. However, this difference was not statistically significant (Studentʼs t test, p = 0.12) (Supplementary Fig. S11). Furthermore, CD31 staining could not clearly confirm decreased angiogenesis within the tumor mass of D492HER2 with KD of ECM1 compared with D492HER2 ctrl. Interestingly though, both ctrl and kdECM1 tumors did show similar ECM1 protein expression levels although ECM1 KD had been confirmed by RT-qPCR prior injection into mice.
Triple-negative (TN) MDA-MB-231 cells also show high ECM1 expression and are capable of increasing endothelial network formation and inducing endothelial feedback
When analyzing the correlation of ECM1 and prognosis, we noted that high expression of ECM1 was not only correlated to worse survival in HER2+ breast cancer patients but also in triple-negative (TN) and generally ER− tumor patients (Supplementary Figs. S4 and S8B). In addition, correlation analysis using the Metabric discovery dataset showed a significant positive correlation between ECM1 expression and CD31 as established endothelial marker in ER− breast cancer (Supplementary Fig. S8E). Therefore, we decided to include MDA-MB-231 as TN and MCF-7 as ER+ cell line in our study. Interestingly, CM from MDA-MB-231 but not MCF-7 also increased endothelial network formation after 4, 24, and 48 h (Supplementary Figs. S8C, D, S9). Similar to D492HER2, MDA-MB-231 expresses high levels of ECM1 in contrast to MCF-7 (Supplementary Fig. S8A). Treating ECs with rECM1 also induced an endothelial feedback promoting MDA-MB-231 migration, as seen for ECM1-expressing D492HER2 (Supplementary Fig. S8F, G). Upon transient ECM1 KD in MDA-MB-231 (RT-qPCR confirmation of ECM1 KD see Supplementary Fig. S8H), its ability to increase endothelial network formation was significantly decreased (Supplementary Fig. S8I). This suggests that ECM1 might play a role in cancer progression through endothelial cross talk in a broader range of breast cancer subtypes than just HER2+.
ECM1 upregulates endothelial NOTCH1 and NOTCH3
To investigate whether treatment with D492HER2-CM upregulates commonly known angiogenesis modulators in ECs, we examined gene expression levels of endothelial VEGFa, VEGFR2 and PDGFb. However, none of these markers showed increased expression upon treatment with D492HER2-CM (Supplementary Fig. S7A).
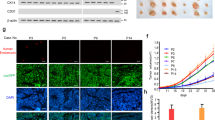
In order to determine transcriptional changes occurring in ECs specifically in response to cancer-secreted ECM1, we performed RNA microarray analysis of ECs treated with CM from D492HER2 and D492HER2 with ECM1 KD. After p value correction and setting the fold change cutoff to twofold, analysis revealed 18 genes upregulated in ECs treated with D492HER2-CM compared with ECs treated with CM from D492HER2-kdECM1 (Supplementary Fig. S7 and Supplementary Table S2). However, the observed changes in genes expression were rather mild. One of the mildly higher expressed genes in ECs treated with D492HER2-CM was fatty acid-binding protein 4 (FABP4). FABP4 itself could not be confirmed as upregulated compared with untreated ECs, however it led our attention to the NOTCH pathway since its expression is regulated by NOTCH1 [51,52,53]. When investigating the expression of the NOTCH receptors 1–4 in ECs, we observed increased NOTCH1 and NOTCH3 gene and protein expression in ECs treated with D492HER2-CM and rECM1 but not with CM from D492HER2-kdECM1 (Fig. 5a).
a ECM1 increases endothelial NOTCH1 and NOTCH3. Increased NOTCH1 and NOTCH3 expression in HUVECs treated with D492HER2-CM and rECM1 but not D492HER2-kdECM1-CM on gene expression level (left) and protein expression level (IF staining, scale bar = 50 µm). Studentʼs t test, ***p < 0.001, **p < 0.01, *p < 0.05, <0.1. b NOTCH inhibition blocks ECM1-induced increase of endothelial network formation. Top: HUVEC endothelial network in H14 (ctrl), 60 ng/ml rECM1, 60 ng/ml rECM1 + DMSO and 60 ng/ml rECM1 + DAPT (20 µM) after 4 h. 4–6 wells as replicates per condition and 1–2 images taken per well at ×10 magnification (scale bar = 100 µm). Bottom: corresponding angiogenesis images (ImageJ angiogenesis analyzer). c Endothelial network formation—quantification. Quantification of master junctions, master segments, total master segment length, and meshes for each image using angiogenesis analyzer plugin. One-way ANOVA, ***p < 0.001, **p < 0.01, *p < 0.05, <0.1. d Endothelial feedback reduction—schematic overview. Schematic workflow of treating ECs using 60 ng/ml rECM1 and blocking NOTCH signaling using DAPT (20 µM). e NOTCH inhibition blocks rECM1-induced endothelial feedback on migration and invasion. Transwell-migration and invasion assays of D492HER2 treated with unconditioned medium, unconditioned HUVEC-CM, DMSO-treated HUVEC-CM, DAPT-treated HUVEC-CM, 60 ng rECM1-treated HUVEC-CM, 60 ng rECM1 + DMSO-treated HUVEC-CM, and 60 ng rECM1 + DAPT-treated HUVEC-CM (NOTCH inhibition). Number of migratory/invasive cells for different treatments (Studentʼs t test, ***p < 0.001, **p < 0.01, *p < 0.05, <0.1).
Blocking endothelial NOTCH signaling interrupts the increase in network formation and feedback induction
Next, we wanted to investigate whether endothelial NOTCH signaling was implicated in the observed increase in endothelial network formation and the induction of an endothelial feedback promoting cancer cell migration and invasion. By blocking endothelial NOTCH using gamma-secretase inhibitor DAPT, interruption in both the increase in network formation and the feedback induction was observed. While treatment with rECM1 resulted in significantly increased numbers of master junctions, master segments, total master segment length, and number of meshes, this effect was no longer visible upon addition of DAPT (Fig. 5b, c). Compared with addition of rECM1 + DMSO, addition of rECM1 + DAPT significantly reduced numbers of master junctions (p = 0.0004), master segments (p = 0.0014), total master segment length (p = 0.0019), and number of meshes (p = 0.0012). Furthermore, CM of rECM1-induced ECs stimulated an increase in cancer cell migration and invasion whereas the addition of DAPT inhibited the induction of this feedback (Fig. 5d, e). Blocking endothelial NOTCH signaling therefore appears to interrupt both cancer-increased network formation as well as feedback induction. Interestingly, addition of DAPT alone to HUVEC did not affect the endothelial network (Supplementary Fig. S12) and feedback on migration and invasion (Fig. 5e). This suggests that blocking endothelial NOTCH by DAPT reduces network formation and feedback only upon elevated NOTCH signaling conditions.
Discussion
In this study, we have demonstrated that the HER2-overexpressing breast cancer cell line, D492HER2, stimulates endothelial network formation in vitro and is capable of inducing an endothelial feedback promoting cancer cell migration toward the endothelium and invasion. We have further identified ECM1 as a secreted factor in D492HER2 that plays a role in mediating both the pro-angiogenic effect of D492HER2 and the feedback induction. rECM1 increases endothelial network formation and induces the feedback while KD of ECM1 interrupts both. Furthermore, D492HER2 no longer show an increased migrative phenotype toward ECs upon ECM1 KD. However, the fact that we observed only a marginal reduction of tumor volume upon ECM1 KD indicates that it may be necessary to knockout ECM1 for in vivo studies and further to evaluate angiogenesis of the tumor-surrounding stroma. This is supported by the study of Wu and colleagues that have performed in vivo studies with ECM1 knockout rather than KD cells [54]. Interestingly, ECM1 protein expression levels were found to be similar in both ctrl and kdECM1 tumors although ECM1 KD had been confirmed by RT-qPCR prior injection into mice. Rather than suggesting no difference in tumor formation between ctrl and kdECM1 cells, these results are a possible indication that a positive selection of ECM1+ cells might have occurred in mice injected with kdECM1 cells. Therefore, our results do not necessarily stand in conflict to previously published data on ECM1 promoting tumor and metastasis formation in vivo [36,37,38, 54, 55]. Indeed, new studies could show that injection of equal amounts of different clones of the same breast cancer cell line MDA-MB-231 leads to formation of tumors with very few cells of one but many cells from another clone [56]. Tracing ECM1+ and ECM1− clones in vivo would offer a great approach to further investigate the occurrence of positive selection of ECM1+ clones and clarify the role of ECM1 during tumor formation.
ECM1 is a secreted glycoprotein and was first identified in the mouse osteogenic stromal cell line MN7 [47]. It was initially described as modulator of proliferation and differentiation of epidermal keratinocytes and basement membrane reconstitution in the skin [48]. However, it has also been linked to angiogenesis and malignant transformation [32,33,34, 37, 38, 48,49,50]. Han et al. demonstrated that rECM1 stimulated proliferation and blood vessel formation in the chorioallantoic membrane of chicken embryos [32] and Wang et al. found it to be expressed particularly in highly malignant breast epithelial tumors [34]. Recently, Wu et al. linked ECM1 to breast cancer progression by suggesting a role of ECM1 in facilitating metastasis through increased invasion and adhesion capacity of ECM1-expressing cancer cells [54]. These data are in line with our findings in the current study. Ferraro et al. recently showed that ECs are capable of increasing migration and invasion of SKBR3 breast cancer cells [57]. However, a direct involvement of cancer-secreted ECM1 in cancer progression through induction of an endothelial feedback supporting cancer migration and invasion has not been reported yet. Here, we show that the increased migration is directed toward the endothelium, suggesting that ECM1 may play a mediating role in inducing ECs to promote cancer cell migration from the primary tumor toward the vasculature and therefore support metastasis formation and tumor progression.
Survival analysis shows that expression of ECM1 in HER2+ breast tumors is associated with significantly reduced distant metastasis-free survival (DMFS). Therefore, targeting ECM1 in tumors overexpressing HER2 could lead to better patient prognosis. This is supported by a study of Lee et al., which showed that ECM1 regulates resistance to trastuzumab, a recombinant antibody targeting HER2 and the most common drug used in treatment of HER2+ breast cancer [49]. Interestingly, the TN breast cancer cell line MDA-MB-231 also expresses high levels of ECM1 and is capable of inducing endothelial network formation in the present study and previous studies [18, 19, 54]. KD of ECM1 in MDA-MB-231 inhibits these pro-angiogenic effects, supporting the mediating role of ECM1 in increasing the endothelial network. We also report that rECM1 is capable of inducing a positive endothelial feedback on MDA-MB-231 migration. Since expression of ECM1 also correlates with worse outcome in TN and generally ER− breast cancer, its molecular involvement in breast cancer progression might be more general rather than only limited to HER2+ cancer. Silencing of ECM1 in MDA-MB-231 led to a decrease in cancer cell migration and invasion capacity, linking its expression to increased tumor progression aggressiveness [54, 55]. Therefore, ECM1 might represent a possible therapeutic target for a broader range of breast cancer patients than just HER2+ individuals. However, the specific relationship between HER2 overexpression and ECM1 still needs to be elucidated. ECM1 expression is upregulated in several HER2+ breast cancer cell lines including ZR7530, SUM44PE, and HCC202. However, other HER2+ cell lines such as SKBR3 or BT474 show no ECM1 upregulation. However, among the breast cancer cell lines with strong ECM1 upregulation according to the GOBO database there are numerous ER− cell lines (e.g., HCC1143, HBL100, SUM1315MO2, MDA-MB-435, and MDA-MB-231). Lee et al. report that high ECM1 expression in HER2+ breast cancer is correlated with poor prognosis and trastuzumab resistance [49]. They suggest that ECM1 may interact with EGFR family receptors that interact with HER2, attenuating therapeutic action by trastuzumab. However, the mechanisms of ECM1 expression regulation itself are not very well understood in HER2+ breast cancer. Smits et al. identified conserved potential binding sites for transcription factors of the Ap1, Ets, and Sp1 family in the human ECM1 gene in MN7 cells [58]. Ye et al. provide evidence that ECM1 is a target of miR-486-3p in cervical cancer. Overexpression of miR-486-3p thereby inhibited cell growth and metastasis by targeting ECM1 [59]. However, we could not detect differential expression of miR-486-3p between D492, D492M, and D492HER2, suggesting that ECM1 might be regulated through a different mechanism in our HER2+ breast cancer system. Generally, investigating the involvement of epigenetic regulation in modulating ECM1 expression and downstream signaling could be of great interest though as it could potentially give novel insights also in the context of cellular cross talk.
In our model, ECM1 was identified as cancer-secreted protein with pro-angiogenic properties that is capable of inducing an endothelial feedback promoting cancer cell migration toward ECs and invasion (Fig. 6). However, the molecular mechanism through which ECM1 mediates this feedback of the vascular niche on cancer progression has not been described previously. Here, we identified an upregulation of NOTCH1 and NOTCH3 receptor expression in ECs upon treatment with D492HER2-CM and rECM1 but not CM from D492HER2 with ECM1 KD. However, the initial microarray candidate, FABP4, could not be confirmed in further experiments, indicating that the subsequent downstream signaling may not involve FABP4. This would coincide with the findings that in addition to NOTCH1, also VEGFa, bFGF, and FOXO1 play potential roles in regulation of FABP4 expression [60]. So far, ECM1 has been associated with EGF signaling [49], and ITGB4/FAK/SOX2/HIF1a [36] and WNT1/ß-catenin [37] pathways, but a mediating role in NOTCH signaling has not been described. NOTCH signaling is very complex and plays a role in a large number of biological processes [61, 62]. Nevertheless, recent studies suggest NOTCH1 as a key player in inducing tumor angiogenesis and therefore promoting tumor progression in cutaneous melanoma and myeloma [25, 26]. Kalucka et al. have shown that NOTCH1 is involved in endothelial barrier function and angiogenesis by controlling fatty acid metabolism [53]. These data would support a possible involvement of endothelial NOTCH in mediating the increase in endothelial network and possibly the induction of the pro-cancerous feedback in breast cancer. Indeed, upon inhibition of endothelial NOTCH signaling, the enhancing effect of ECM1 on network formation and feedback was no longer observed, indicating the existence of a ECM1/NOTCH axis involved in mediating the cross talk between cancer cells and endothelium in terms of promoting cancer progression (Fig. 6).
ECM1 secreted by HER2+, TN, or ER− breast cancer cells increases endothelial network formation and induces endothelial feedback promoting cancer cell migration and invasion through upregulation of NOTCH1 and NOTCH3 expression. Knockdown of ECM1 in cancer cells and NOTCH inhibition in ECs inhibits the network formation increase and the feedback induction.
In conclusion, we have identified ECM1 as a cancer-secreted pro-angiogenic factor involved in inducing the endothelial niche to support cancer progression through increased cancer cell migration and invasion. Moreover, our data indicate a novel, mediating role of ECM1 in endothelial NOTCH signaling, which appears to be involved in mediating the induction of the cancer-promoting endothelial feedback (Fig. 6). Taken together, our findings might open up new possibilities for cancer treatment by understanding how an activated vascular niche provides positive feedback to the cancer and targeting this interaction between cancer cells and their microenvironment.
References
Hassiotou F, Geddes D. Anatomy of the human mammary gland: current status of knowledge. Clinical Anat. 2013;26:29–48.
Sigurdsson V, Fridriksdottir AJ, Kjartansson J, Jonasson JG, Steinarsdottir M, Petersen OW, et al. Human breast microvascular endothelial cells retain phenotypic traits in long-term finite life span culture. In Vitro Cell Dev Biol Anim. 2006;42:332–40.
Ingthorsson S, Sigurdsson V, Fridriksdottir A Jr., Jonasson JG, Kjartansson J, Magnusson MK, et al. Endothelial cells stimulate growth of normal and cancerous breast epithelial cells in 3D culture. BMC Res Notes. 2010;3:184.
Gudjonsson T, Ronnov-Jessen L, Villadsen R, Bissell MJ, Petersen OW. To create the correct microenvironment: three-dimensional heterotypic collagen assays for human breast epithelial morphogenesis and neoplasia. Methods. 2003;30:247–55.
Bergthorsson JT, Magnusson MK, Gudjonsson T. Endothelial-rich microenvironment supports growth and branching morphogenesis of prostate epithelial cells. Prostate. 2013;73:884–96.
Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol. 1982;99:31–68.
Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, et al. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol. 2013;15:807–17.
Erler JT, Weaver VM. Three-dimensional context regulation of metastasis. Clin Exp Metastasis. 2009;26:35–49.
Hoye AM, Erler JT. Structural ECM components in the premetastatic and metastatic niche. Am J Physiol Cell Physiol. 2016;310:C955–67.
Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–22.
Oskarsson T, Batlle E, Massague J. Metastatic stem cells: sources, niches, and vital pathways. Cell Stem Cell. 2014;14:306–21.
Pollard JW. Macrophages define the invasive microenvironment in breast cancer. J Leukoc Biol. 2008;84:623–30.
Komohara Y, Jinushi M, Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 2014;105:1–8.
Komohara Y, Takeya M. CAFs and TAMs: maestros of the tumour microenvironment. J Pathol. 2017;241:313–5.
Shan T, Chen S, Chen X, Lin WR, Li W, Ma J, et al. Cancer-associated fibroblasts enhance pancreatic cancer cell invasion by remodeling the metabolic conversion mechanism. Oncol Rep. 2017;37:1971–9.
Zhang Z, Li X, Sun W, Yue S, Yang J, Li J, et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017;397:33–42.
Ghajar CM. Metastasis prevention by targeting the dormant niche. Nat Rev Cancer. 2015;15:238–47.
Lee E, Fertig EJ, Jin K, Sukumar S, Pandey NB, Popel AS. Breast cancer cells condition lymphatic endothelial cells within pre-metastatic niches to promote metastasis. Nat Commun. 2014;5:4715.
Lee E, Pandey NB, Popel AS. Crosstalk between cancer cells and blood endothelial and lymphatic endothelial cells in tumour and organ microenvironment. Expert Rev Mol Med. 2015;17:e3.
Lee E, Pandey NB, Popel AS. Lymphatic endothelial cells support tumor growth in breast cancer. Sci Rep. 2014;4:5853.
Franses JW, Baker AB, Chitalia VC, Edelman ER. Stromal endothelial cells directly influence cancer progression. Sci Transl Med. 2011;3:66ra5.
Franses JW, Drosu NC, Gibson WJ, Chitalia VC, Edelman ER. Dysfunctional endothelial cells directly stimulate cancer inflammation and metastasis. Int J Cancer. 2013;133:1334–44.
Ucuzian AA, Gassman AA, East AT, Greisler HP. Molecular mediators of angiogenesis. J Burn Care Res. 2010;31:158–75.
Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1alpha) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol Cell Biol. 2001;21:3995–4004.
Murtas D, Piras F, Minerba L, Maxia C, Ferreli C, Demurtas P, et al. Activated Notch1 expression is associated with angiogenesis in cutaneous melanoma. Clin Exp Med. 2015;15:351–60.
Guo D, Li C, Teng Q, Sun Z, Li Y, Zhang C. Notch1 overexpression promotes cell growth and tumor angiogenesis in myeloma. Neoplasma. 2013;60:33–40.
Sigurdsson V, Hilmarsdottir B, Sigmundsdottir H, Fridriksdottir AJ, Ringner M, Villadsen R, et al. Endothelial induced EMT in breast epithelial cells with stem cell properties. PLoS ONE. 2011;6:e23833.
Gudjonsson T, Villadsen R, Nielsen HL, Ronnov-Jessen L, Bissell MJ, Petersen OW. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev. 2002;16:693–706.
Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, et al. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101.
Briem E, Ingthorsson S, Traustadottir GA, Hilmarsdottir B, Gudjonsson T. Application of the D492 cell lines to explore breast morphogenesis, EMT and cancer progression in 3D culture. J Mammary Gland Biol Neoplasia. 2019;24:139–47.
Ingthorsson S, Andersen K, Hilmarsdottir B, Maelandsmo GM, Magnusson MK, Gudjonsson T. HER2 induced EMT and tumorigenicity in breast epithelial progenitor cells is inhibited by coexpression of EGFR. Oncogene. 2015;35:4244–55.
Han Z, Ni J, Smits P, Underhill CB, Xie B, Chen Y, et al. Extracellular matrix protein 1 (ECM1) has angiogenic properties and is expressed by breast tumor cells. FASEB J. 2001;15:988–94.
Sercu S, Zhang L, Merregaert J. The extracellular matrix protein 1: its molecular interaction and implication in tumor progression. Cancer Invest. 2008;26:375–84.
Wang L, Yu J, Ni J, Xu X-M, Wang J, Ning H, et al. Extracellular matrix protein 1 (ECM1) is over-expressed in malignant epithelial tumors. Cancer Lett. 2003;200:57–67.
Chen H, Jia W, Li J. ECM1 promotes migration and invasion of hepatocellular carcinoma by inducing epithelial-mesenchymal transition. World J Surg Oncol. 2016;14:195.
Gan L, Meng J, Xu M, Liu M, Qi Y, Tan C, et al. Extracellular matrix protein 1 promotes cell metastasis and glucose metabolism by inducing integrin beta4/FAK/SOX2/HIF-1alpha signaling pathway in gastric cancer. Oncogene. 2017;37:744–55.
Lee KM, Nam K, Oh S, Lim J, Kim RK, Shim D, et al. ECM1 regulates tumor metastasis and CSC-like property through stabilization of beta-catenin. Oncogene. 2015;34:6055–65.
Wang Z, Zhou Q, Li A, Huang W, Cai Z, Chen W. Extracellular matrix protein 1 (ECM1) is associated with carcinogenesis potential of human bladder cancer. Onco Targets Ther. 2019;12:1423–32.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22.
Guillen J. FELASA guidelines and recommendations. J Am Assoc Lab Anim Sci. 2012;51:311–21.
Rosenblat JD, Brietzke E, Mansur RB, Maruschak NA, Lee Y, McIntyre RS. Inflammation as a neurobiological substrate of cognitive impairment in bipolar disorder: evidence, pathophysiology and treatment implications. J Affect Disord. 2015;188:149–59.
Kastrup J. Can YKL-40 be a new inflammatory biomarker in cardiovascular disease? Immunobiology. 2012;217:483–91.
Vignon E. Is glycoprotein YKL40 a new marker for joint disorders? Joint Bone Spine. 2001;68:454–6.
Zhou Y, Peng H, Sun H, Peng X, Tang C, Gan Y, et al. Chitinase 3-like 1 suppresses injury and promotes fibroproliferative responses in Mammalian lung fibrosis. Sci Transl Med. 2014;6:240ra76.
Francescone RA, Scully S, Faibish M, Taylor SL, Oh D, Moral L, et al. Role of YKL-40 in the angiogenesis, radioresistance, and progression of glioblastoma. J Biol Chem. 2011;286:15332–43.
Shao R. YKL-40 acts as an angiogenic factor to promote tumor angiogenesis. Front Physiol. 2013;4:122.
Mathieu E, Meheus L, Raymackers J, Merregaert J. Characterization of the osteogenic stromal cell line MN7: identification of secreted MN7 proteins using two-dimensional i’olyacrylamide gel electrophoresis, western blotting, and microsequencing. J Bone Miner Res. 1994;9:903–13.
Oyama N, Merregaert J. The extracellular matrix protein 1 (ECM1) in molecular-based skin biology. In: Farage M., Miller K., Maibach H. (eds) Textbook of Aging Skin. Springer, Berlin, Heidelberg. 2017. p. 91–110.
Lee KM, Nam K, Oh S, Lim J, Kim YP, Lee JW, et al. Extracellular matrix protein 1 regulates cell proliferation and trastuzumab resistance through activation of epidermal growth factor signaling. Breast Cancer Res. 2014;16:479.
Sercu S, Zhang M, Oyama N, Hansen U, Ghalbzouri AE, Jun G, et al. Interaction of extracellular matrix protein 1 with extracellular matrix components: ECM1 is a basement membrane protein of the skin. J Invest Dermatol. 2008;128:1397–408.
Harjes U, Bridges E, Gharpure KM, Roxanis I, Sheldon H, Miranda F, et al. Antiangiogenic and tumour inhibitory effects of downregulating tumour endothelial FABP4. Oncogene. 2017;36:912–21.
Bruning U, Morales-Rodriguez F, Kalucka J, Goveia J, Taverna F, Queiroz KCS, et al. Impairment of angiogenesis by fatty acid synthase inhibition involves mTOR malonylation. Cell Metab. 2018;28:866.e15–80.e15.
Kalucka J, Bierhansl L, Conchinha NV, Missiaen R, Elia I, Bruning U, et al. Quiescent endothelial cells upregulate fatty acid beta-oxidation for vasculoprotection via redox homeostasis. Cell Metab. 2018;28:881.e13–94.e13.
Wu Q, Chen D, Luo Q, Yang Q, Zhao C, Zhang D, et al. Extracellular matrix protein 1 recruits moesin to facilitate invadopodia formation and breast cancer metastasis. Cancer Lett. 2018;437:44–55.
Gomez-Contreras P, Ramiro-Diaz JM, Sierra A, Stipp C, Domann FE, Weigel RJ, et al. Extracellular matrix 1 (ECM1) regulates the actin cytoskeletal architecture of aggressive breast cancer cells in part via S100A4 and Rho-family GTPases. Clin Exp Metastasis. 2017;34:37–49.
Martin-Pardillos A, Valls Chiva A, Bande Vargas G, Hurtado Blanco P, Pineiro Cid R, Guijarro PJ, et al. The role of clonal communication and heterogeneity in breast cancer. BMC Cancer. 2019;19:666.
Ferraro DA, Patella F, Zanivan S, Donato C, Aceto N, Giannotta M, et al. Endothelial cell-derived nidogen-1 inhibits migration of SK-BR-3 breast cancer cells. BMC Cancer. 2019;19:312.
Smits P, Bhalerao J, Merregaert J. Molecular cloning and characterization of the mouse Ecm1 gene and its 5’ regulatory sequences. Gene. 1999;226:253–61.
Ye H, Yu X, Xia J, Tang X, Tang L, Chen F. MiR-486-3p targeting ECM1 represses cell proliferation and metastasis in cervical cancer. Biomed Pharmacother. 2016;80:109–14.
Furuhashi M, Saitoh S, Shimamoto K, Miura T. Fatty acid-binding protein 4 (FABP4): pathophysiological insights and potent clinical biomarker of metabolic and cardiovascular diseases. Clin Med Insights Cardiol. 2014;8(Suppl 3):23–33.
Traustadottir GA, Jensen CH, Thomassen M, Beck HC, Mortensen SB, Laborda J, et al. Evidence of non-canonical NOTCH signaling: delta-like 1 homolog (DLK1) directly interacts with the NOTCH1 receptor in mammals. Cell Signal. 2016;28:246–54.
Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6.
Acknowledgements
We thank the FingerPrints Proteomics Facility at the University of Dundee for performing the mass spectrometry and the Center for Systems Biology at the University of Iceland for the technical support during mass spectrometry data analysis. Further, we want to thank the HI-STEM laboratory and the sequencing core facility at the German Cancer Research Center, DKFZ Heidelberg, Germany, for technical support and performing the RNA microarray. Finally, we thank Prof. Dr Haraldur Haraldsson, University of Iceland for providing primary human umbilical vascular ECs. Human umbilical vascular ECs were obtained from umbilical chords at the childbirth department, University Hospital Iceland (ethical application nr. 35/2013). This work was supported by Grants from Landspitali University Hospital Science Fund, University of Iceland Research Fund (Grant of Excellence: #152144051 Doctoral Grant: #185042051), and Icelandic Science and Technology Policy.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest. The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Steinhaeuser, S.S., Morera, E., Budkova, Z. et al. ECM1 secreted by HER2-overexpressing breast cancer cells promotes formation of a vascular niche accelerating cancer cell migration and invasion. Lab Invest 100, 928–944 (2020). https://doi.org/10.1038/s41374-020-0415-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-020-0415-6
This article is cited by
-
TET2-mediated ECM1 hypomethylation promotes the neovascularization in active proliferative diabetic retinopathy
Clinical Epigenetics (2024)
-
Truncated O-glycosylation in metastatic triple-negative breast cancer reveals a gene expression signature associated with extracellular matrix and proteolysis
Scientific Reports (2024)
-
Presence of crown-like structures in breast adipose tissue; differences between healthy controls, BRCA1/2 gene mutation carriers and breast cancer patients
Breast Cancer Research and Treatment (2024)
-
Extracellular matrix protein 1 (ECM1) is a potential biomarker in B cell acute lymphoblastic leukemia
Clinical and Experimental Medicine (2024)