Abstract
Hypoxia is an important feature of the tumor microenvironment and is associated with glioma progression and patient outcome. Exosomes have been implicated in the intercellular communication in the tumor microenvironment. However, the effects of hypoxic glioma exosomes on glioma migration and invasion and the underlying mechanisms remain poorly understood. In this study, we found that exosomes derived from hypoxic glioma cells (H-GDEs) promoted normoxic glioma migration and invasion in vitro and in vivo. Given that exosomes can regulate recipient cell functions by delivering microRNAs, we further revealed miR-1246 and miR-10b-5p were upregulated significantly in H-GDEs and delivered to normoxic glioma cells by H-GDEs. Moreover, we determined the clinical relevance of miR-1246 and miR-10b-5p in glioma patients. Subsequent investigations indicated that miR-1246 and miR-10b-5p markedly induced glioma migration and invasion in vitro and in vivo. Finally, we demonstrated that miR-1246 and miR-10b-5p induced glioma migration and invasion by directly targeting FRK and TFAP2A respectively. In conclusion, our findings suggest that the hypoxic microenvironment stimulates glioma to generate miR-1246- and miR-10b-5p-rich exosomes that are delivered to normoxic glioma cells to promote their migration and invasion; treatment targeting miR-1246 and miR-10b-5p may impair the motility of gliomas, providing a novel direction for the development of antitumor therapy.
Similar content being viewed by others
Introduction
Glioma (especially glioblastoma multiforme [GBM]), with distinct characteristics, is the most common and malignant primary tumor of the central nervous system in adults [1]. Despite advances in surgical techniques, radiotherapy, chemotherapy, and therapies targeting mechanisms intrinsic to gliomas, the prognosis of GBM patients remains poor, with a median survival time of less than 15 months following primary diagnosis [1]. The recalcitrance of malignant gliomas to standard surgical therapy and other therapy strategies is due to the ability of gliomas to invade diffusely into brain parenchyma and the unique tumor microenvironment, which can facilitate glioma progression [2, 3].
Hypoxia, a well-characterized feature of the tumor microenvironment [4], is a potent driving force for tumor progression [5]. The effects of hypoxia on tumors include selection of more aggressive tumor cells [6], switches in metabolism [7], and enhanced angiogenesis [8]. Furthermore, numerous studies have demonstrated that hypoxia promotes the epithelial-to-mesenchymal transition (EMT) [5], tumor invasiveness [9], and metastasis [10], suggesting an important role for hypoxia in tumor metastasis.
Exosomes are small membrane vesicles (30–100 nm in size) that can be secreted by, e.g., immune cells and tumor cells [11]. As an important component of the tumor microenvironment, exosomes, which contain multiple contents (e.g., mRNAs, proteins and non-coding RNAs), are crucial in intercellular communication by delivering their contents [12]. Tumor exosomes have been demonstrated to facilitate tumor progression by modulating tumor cells and surrounding stromal cells [13, 14], reprogramming and suppressing immune cells [15, 16], and establishing pre-metastatic sites [17]. Moreover, a recent study has revealed that hypoxia can enhance the release and alter the content of exosomes [18], thereby influencing intercellular communication in the tumor microenvironment [19]. We have demonstrated that through regulating the content of glioma-derived exosomes (GDEs), hypoxia can promote the expansion and induce immunosuppressive function of myeloid-derived suppressor cells [20]. These findings indicate that hypoxia may facilitate tumor progression and improve the tumor microenvironment, at least in part, through its effect on exosome release and content. Hence, we investigated whether hypoxic glioma-derived exosomes (H-GDEs) can serve as communicators between hypoxic and normoxic glioma cells through transporting microRNAs, mediating the promotion of normoxic glioma migration and invasion by the hypoxic microenvironment.
In the present study, we performed exosome identification and determined that H-GDEs could be internalized by glioma cells. Furthermore, we demonstrated that compared with exosomes derived from normoxic glioma cells (N-GDEs), H-GDEs could increase the migration and invasion abilities of normoxic glioma cells. Next, we found miR-1246 and miR-10b-5p were increased significantly by hypoxia, exhibiting high expression levels in H-GDEs. We also determined the clinical relevance of miR-1246 and miR-10b-5p in glioma patients. Moreover, miR-1246 and miR-10b-5p showed a strong ability to promote glioma migration and invasion. Finally, we found that miR-1246 and miR-10b-5p increased glioma cell motility by directly targeting FRK and TFAP2A respectively.
Materials and methods
Cell lines and hypoxia treatment
U87MG, U251, A172 glioma cell lines and the HEK293T cell line were obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China). The primary human glioblastoma cell line (GBM#P3) was provided by the Department of Biomedicine at the University of Bergen (Norway). U87MG, U251, A172, and HEK293T cell lines were cultured in DMEM (Thermo Fisher Scientific, USA) supplemented with 10% FBS (Thermo Fisher Scientific). GBM#P3 cells were cultured as previously described [20]. The cell lines were authenticated by short tandem repeat (STR) profiling and identified without contamination before the study started. The cells were cultured under 20% O2 or 1% O2 conditions balanced with N2.
Exosome isolation and labeling
Exosomes were isolated from the cell culture supernatant collected from glioma cell lines as previously described [21]. Briefly, the supernatant was centrifuged at 300 g for 10 min, 2000 g for 10 min, and 10,000 g for 30 min, followed by filtration through a 0.22 μm filter. Then, the supernatant was ultracentrifuged at 100,000 g for 70 min, followed by a washing step of the exosome pellet with PBS at 100,000 g for 70 min (Ultracentrifuge, Beckman Coulter). The exosome pellet was resuspended in PBS and stored at –80 °C. Purified exosomes were labeled with the green fluorescent linker PKH67 (Sigma-Aldrich, USA) as previously described [20, 21]. Briefly, the exosomes diluted in PBS were added to 1 ml Diluent C (Sigma-Aldrich). In parallel, 4 μl PKH67 dye was added to 1 ml Diluent C and incubated with the exosome solution for 4 min. To bind excess dye, 2 ml 0.5% BSA was added. The labeled exosomes were washed at 100,000 g for 1 h, and the exosome pellet was diluted in PBS and used for uptake experiments.
Transmission electron microscopy (TEM) and qNano analysis
Isolated exosomes were photographed using TEM as previously described [20]. Briefly, purified exosomes were loaded onto a carbon-coated electron microscopy grid and examined using TEM. Glutaraldehyde (3%) was placed on the grids for 5 min. Then the grids were washed by tri-distilled water for 2 min and repeated ten times. Next, the grids were incubated with uranyl-acetate solution (4%) for 10 min and methylcellulose solution (1%) for 5 min. At last, grids were dried and observed using a TEM. qNano (Izon Sciences Ltd, NZ) analysis was used to measure the exosome particle size and concentration [20].
Western blot
Proteins were extracted from GDEs and glioma cells. The following antibodies were used: anti-calnexin (Abcam, UK, ab133615), anti-TSG101 (Abcam, ab125011), anti-N-cadherin (Abcam, ab76011), anti-MMP2 (Abcam, ab92536), anti-MMP9 (Abcam, ab38898), anti-Vimentin (Abcam, ab92547), anti-GAPDH (Cell Signaling Technology, USA, 5174), anti-FRK (Abcam, ab91623), anti-TFAP2A (Cell Signaling Technology, 3215).
Migration and invasion assay
Migration and invasion assays were conducted with uncoated and matrigel-coated (Corning Costar, USA) transwell chambers (Corning Costar) according to the manufacturer’s instructions. After treatment, glioma cells were seeded into the top chamber in DMEM with 1% FBS and DMEM with 30% FBS was filled into the bottom chamber. Glioma cells that migrated to or invaded the bottom of the membrane were fixed and stained.
The 3D spheroid invasion assay was conducted with spheroid formation matrix and invasion matrix (Trevigen, USA) according to the manufacturer’s instructions. Glioma cells were incubated with spheroid formation matrix for 24–72 h to generate spheroids. Then spheroids were embedded into invasion matrix with or without treatment. Glioma spheroids were photographed every 24 h by a light microscope (Leica, German).
ELISA
Cell culture medium was collected 72 h after the indicated treatment. The secretion of MMP2 was detected by ELISA (Proteintech, USA) according to the manufacturer’s instructions.
Hematoxylin and eosin (HE) staining and immunohistochemistry (IHC) staining
HE and IHC staining were performed as previously described [22]. The following primary antibodies: anti-N-cadherin (Servicebio, China), anti-MMP2 (Servicebio) and anti-Ki67 (Servicebio) were used for IHC staining.
RNA extraction and qRT-PCR
For exosome RNA extraction, exosomes were firstly isolated by using Exoquick (System Biosciences). Then exosome RNA was extracted by using the SeraMirTM Exosome RNA Extraction Kit (System Biosciences, USA).
TRIzol (Thermo Fisher Scientific) was used for the total cellular RNA extraction. Reverse transcription and quantitative RT-PCR were performed as previously described [22]. Primer sequences are presented in Supplementary Table S1.
MicroRNA mimics, small interfering RNA, plasmids and virus transfection
Control microRNAs, miR-1246 mimics, miR-10b-5p mimics, control siRNAs, FRK siRNAs, and TFAP2A siRNAs were purchased from GenePharma (China). The plasmids used in this study were pcDNA3.1-FRK, pcDNA3.1-TFAP2A, pGL3-FRK-3′UTR-wild, pGL3-FRK-3′UTR-mut, pGL3-TFAP2A-3′UTR-wild, pGL3-TFAP2A-3′UTR-mut (Bio-Asia, China). MiR-1246, miR-10b-5p overexpression and control lentiviruses were synthesized by Genechem (China). Transfection of microRNA mimics, siRNAs and plasmids was performed using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. U87MG and U251 glioma cells were transfected with miR-1246 or miR-10b-5p overexpression lentiviruses for 72 h according to the manufacturer’s instructions. Sequences of microRNA mimics and siRNAs are presented in Supplementary Table S2. The transfection efficiencies of siRNAs and viruses were determined (Supplementary Figs. S1 and S2).
Luciferase reporter assay
The online microRNA prediction tools TargetScan and miRDB were used to predict the targets of miR-1246 and miR-10b-5p. The reporter plasmids pGL3-FRK-3′UTR-wild, pGL3-FRK-3′UTR-mut, pGL3-TFAP2A-3′UTR-wild or pGL3-TFAP2A-3′UTR-mut were co-transfected with miR-1246 mimics or miR-10b-5p mimics in 293T cells. Two days later, a dual luciferase reporter assay kit (Promega, USA) was used to measure the reporter gene activity according to the manufacturer’s instructions.
Animal studies
Four-week-old male nude mice were purchased from the Model Animal Research Center of Nanjing University (China). For the establishment of xenografts, the luciferase-labeled glioma cells (106 cells per mouse) were injected into mouse brains by using a stereotactic apparatus (KD Scientific, USA). For the N-GDEs and H-GDEs groups, N-GDEs and H-GDEs were injected into the tail vein every 3 days respectively. The IVIS Lumina series III (PerkinElmer, USA) was used for the in vivo bioluminescent imaging every 5 days after implantation. All mice were followed-up on every day and sacrificed when they exhibited brain tumor-related symptoms. Excised tumor tissues were used for HE and IHC staining.
Studies that included mice have been approved by the Animal Care and Use Committee of Qilu Hospital of Shandong University.
Statistical analysis
SPSS 22 was used to perform data analysis and GraphPad Prism 6 was used to realize data visualization. All results are presented as the mean ± SD. Statistical significance was determined by Student’s t test and one-way ANOVA test. Survival curves were estimated by Kaplan–Meier analysis and compared by the log-rank test. P values < 0.05 (denoted by ‘*’) were considered significant; P values < 0.01 (denoted by ‘**’) were considered significant; and P > 0.05 (denoted by ‘N.S.’) was considered not significant.
Results
Identification of glioma exosomes and internalization of glioma exosomes by glioma cells
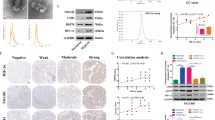
GDEs were isolated from the supernatant of U87, U251, A172 and GBM#P3 glioma cells. TEM images showed the purified exosomes were particles ranging from 30 to 100 nm in diameter that exhibited a rounded morphology (Fig. 1a). The qNano analysis revealed the distribution of exosome size and concentration (Fig. 1b). The presence of the exosomal marker TSG101 and the absence of the negative marker calnexin in isolated GDEs were further confirmed by western blot (Fig. 1c). To investigate whether H-GDEs could be internalized by glioma cells, we incubated glioma cells with H-GDEs labeled with fluorescent PKH67 (a membrane phospholipid dye). As shown by confocal microscopy, the internalization of PKH67-labeled H-GDEs by glioma cells increased over time (Fig. 1d), indicating H-GDEs could deliver their biological content to glioma cells.
a Representative electron micrograph of exosomes isolated from normoxic and hypoxic U87MG, U251, A172 and GBM#P3 conditioned medium. Scale bar, 100 nm. b qNano analysis showing the distribution of size and concentration of exosomes derived from U87MG and GBM#P3 cells. c Western blot analysis of the exosome-positive marker TSG101 and the exosome-negative marker calnexin in N-GDEs, H-GDEs and glioma cells. d Representative confocal microscopy images showing the internalization of PKH67-labeled H-GDEs (green) by normoxic U87MG and U251 glioma cells. Scale bar, 10 μm.
Hypoxic glioma-derived exosomes induce normoxic glioma cell migration and invasion
To explore the effects of H-GDEs on the migration and invasion of normoxic glioma cells, we incubated normoxic glioma cells with N-GDEs and H-GDEs respectively. The migration and invasion assay demonstrated that H-GDEs could significantly promote the migration and invasion abilities of normoxic glioma cells compared with N-GDEs (Fig. 2a, b). Consistent with the migration and invasion assay results, H-GDEs markedly increased the invaded area of normoxic glioma spheroids in the 3D invasion model (Fig. 2c, d). Matrix metalloproteinases (MMPs) can induce extracellular matrix degradation and facilitate tumor invasion [23]. MMP2 and MMP9 are important MMPs in glioma migration and invasion [24]. Moreover, the EMT is crucial in tumor metastasis [25]. Given the importance of MMP2, MMP9, and EMT-related proteins in glioma migration and invasion, we determined whether H-GDEs would affect the expression of these proteins. Western blot analysis revealed that H-GDEs could increase the expression of MMP2, MMP9, N-cadherin, and vimentin compared with N-GDEs (Fig. 2e). Moreover, H-GDEs also induced the secretion of MMP2 in U87MG glioma cells (Supplementary Fig. S3A). Collectively, these results indicate that H-GDEs can promote normoxic glioma migration and invasion in vitro. Next, we determined the effects of H-GDEs on glioma migration and invasion in vivo. To this end, U87MG glioma cells were implanted in nude mice in situ. Then N-GDEs or H-GDEs were injected into the tail vein every 3 days. By in vivo bioluminescent imaging, we found a stronger bioluminescence signal in H-GDE-injected mice compared to N-GDE-injected mice, suggesting H-GDEs induce higher aggressivity of the glioma compared to N-GDEs (Fig. 2f). Furthermore, we collected the xenografts from the mice in N-GDEs and H-GDEs groups for HE staining. The HE staining results revealed that compared with N-GDEs, H-GEDs rendered the xenograft margin irregular and ill-defined, implying that H-GDEs promote glioma infiltration (Fig. 2g). Concordantly, mice in the H-GDEs group succumbed to the brain tumor significantly earlier than those in the N-GDEs group (Fig. 2h). To further substantiate these results, we stained the xenografts by immunohistochemistry (IHC). IHC staining confirmed that H-GDEs increased the expression of N-cadherin, MMP2, and Ki67 (a cell proliferation marker), in agreement with the in vitro western blot results (Fig. 2i, j and Supplementary Fig. S3B). Collectively, these results demonstrate that H-GDEs induce normoxic glioma cell migration and invasion both in vitro and in vivo.
a, b Analysis of glioma cell migration and invasion by transwell assay. a N-GDE- and H-GDE-treated U87MG, U251, and A172 glioma cells that migrated to (without Matrigel coating) or invaded (with Matrigel coating) the bottom surface were stained with crystal violet and observed by light microscopy. Scale bar, 100 μm. b Quantitative analysis of the N-GDE- and H-GDE-treated U87MG, U251, and A172 cells that migrated to or invaded the bottom surface. c Analysis of glioma invasion by 3D invasion assay. Representative images of invaded spheroids in 3D invasion assay for N-GDE- and H-GDE-treated U87MG, U251, and A172 glioma cells. Scale bar, 200 mm. d Quantitative analysis of area covered by N-GDE- and H-GDE-treated invading U87MG, U251, and A172 glioma cells. e Western blot analysis of N-cadherin, MMP2, MMP9, and vimentin in N-GDE- and H-GDE-treated U87MG, U251, and A172 glioma cells. f In vivo bioluminescent imaging of N-GDE- and H-GDE-treated U87MG glioma cells implanted in situ in nude mice. Representative images and quantification (15 days after implantation) are shown (five mice in each group). g HE staining of sections from mouse brain xenografts with N-GDE- and H-GDE-treated U87MG glioma cells. Scale bar, 200 μm. h Survival analysis for nude mice bearing N-GDE- and H-GDE-treated U87MG glioma cell xenografts (P < 0.05 by log-rank analysis). i Quantitative analysis of IHC staining for N-cadherin and MMP2 of sections from mouse brain xenografts with N-GDE- and H-GDE-treated U87MG glioma cells. j Quantitative analysis of IHC staining for Ki67 of sections from mouse brain xenografts with N-GDE- and H-GDE-treated U87MG glioma cells. Data are shown as the mean ± SD of three independent experiments. Statistical significance was determined using Student’s t test and log-rank analysis (*P < 0.05; **P < 0.01).
MiR-1246 and miR-10b-5p are enriched in H-GDEs and have clinical relevance in glioma patients
Exosomes have been demonstrated to regulate recipient cell function by delivering their content [12]. Being the most abundant type of RNA in exosomes [26], microRNAs are believed to play a vital role in exosome-mediated intercellular communication. We have determined the microRNA expression profiles of N-GDEs and H-GDEs by using high-throughput sequencing (miRNA-seq) in our previous study [20]. Here we focused on miR-1246 and miR-10b-5p for further study because (i) both miR-1246 and miR-10b-5p levels were markedly increased in H-GDEs; (ii) miR-1246 and miR-10b-5p were expressed at high levels in H-GDEs; and (iii) some studies have demonstrated that miR-1246 and miR-10b-5p promote tumor metastasis [27, 28], while their functions on glioma migration and invasion have not been determined. To substantiate the miRNA-seq results, we examined miR-1246 and miR-10b-5p expression levels in N-GDEs and H-GDEs using qRT-PCR. Consistent with our miRNA-seq results, the expression level of miR-1246 and miR-10b-5p in H-GDEs was markedly higher than that in N-GDEs (Fig. 3a). In addition, normoxic glioma cells incubated with H-GDEs exhibited higher levels of miR-1246 and miR-10b-5p than those treated with N-GDEs (Fig. 3b), suggesting these two microRNAs could be transferred to recipient glioma cells through H-GDEs. Together with the internalization results of H-GDEs into normoxic glioma cells, these results indicate that miR-1246 and miR-10b-5p could be delivered to normoxic glioma cells by H-GDEs. Moreover, by using the Chinese Glioma Genome Atlas (CGGA) database, we found both miR-1246 and miR-10b-5p expressions were correlated with glioma grades (Fig. 3c, d and Supplementary Fig. S4A, B). Compared with LGGs, GBMs had higher expression levels of miR-1246 and miR-10b-5p, suggesting these microRNAs may be involved in glioma progression. Moreover, both miR-1246 and miR-10b-5p were associated with glioma patients’ survival. Higher expression level of miR-1246 and miR-10b-5p indicated poorer prognosis of glioma patients (Fig. 3e, f and Supplementary Fig. S4C–F).
a Quantitative RT-PCR analysis of miR-1246 and miR-10b-5p expression in N-GDEs and H-GDEs from U87MG and U251 glioma cells. b Quantitative RT-PCR analysis of miR-1246 and miR-10b-5p expression in N-GDE- and H-GDE-treated U87MG and U251 glioma cells. c Expression levels of miR-1246 in LGG and GBM in CGGA database. d Expression levels of miR-10b in LGG and GBM in CGGA database. e Survival analysis for glioma patients with low and high expression of miR-1246. f Survival analysis for glioma patients with low and high expression of miR-10b. Data are shown as the mean ± SD of three independent experiments. Statistical significance was determined using Student’s t test and log-rank analysis (*P < 0.05; **P < 0.01).
MiR-1246 and miR-10b-5p promote glioma cell migration and invasion
Both miR-1246 and miR-10b-5p in GDEs could be upregulated by hypoxia and transferred to recipient glioma cells, which prompted us to investigate their effects on glioma migration and invasion. The migration and invasion assay showed that introduction of both miR-1246 mimics and miR-10b-5p mimics prominently promoted glioma cell migration and invasion (Fig. 4a, b). In addition, miR-1246 and miR-10b-5p also induced hypoxic U87MG glioma cell migration and invasion (Supplementary Fig. S5A, B). Consistent with the migration and invasion assay results, both miR-1246 and miR-10b-5p markedly increased the invaded area of glioma spheroids in the 3D invasion model (Fig. 4c, d). Moreover, western blot analysis demonstrated that miR-1246 and miR-10b-5p resulted in higher expression levels of MMP2, MMP9, N-cadherin and vimentin compared with the control group (Fig. 4e). Furthermore, miR-1246 and miR-10b-5p also induced the secretion of MMP2 in U87MG glioma cells (Supplementary Fig. S5C). Collectively, these results indicate that miR-1246 and miR-10b-5p can promote glioma migration and invasion in vitro. To further confirm our results on miR-1246 and miR-10b-5p function, we performed in vivo experiments using a xenograft tumor mouse model. MiR-1246-overexpressing, miR-10b-5p-overexpressing, or miR-NC U87MG glioma cells were implanted in nude mice in situ. By in vivo bioluminescent imaging, we found that both miR-1246 and miR-10b-5p overexpression could increase glioma aggressivity, as stronger bioluminescence signals were observed compared to the miR-NC group (Fig. 4f). Moreover, we collected the xenografts derived from the mice in miR-1246, miR-10b-5p, and miR-NC groups for HE staining. The HE staining results showed that both miR-1246 and miR-10b-5p rendered the xenograft margin irregular and ill-defined, suggesting that both miR-1246 and miR-10b-5p promote glioma infiltration (Fig. 4g). Concordantly, overexpression of miR-1246 and miR-10b-5p significantly decreased the survival rate of nude mice bearing glioma xenografts compared with the miR-NC group (Fig. 4h). To further extend these findings, we used the xenografts for IHC staining. IHC staining substantiated our results that miR-1246 and miR-10b-5p increased the expression of N-cadherin, MMP2, and Ki67, in accordance with our in vitro western blot results (Fig. 4i, j and Supplementary Fig. S5D). Collectively, these results indicate that miR-1246 and miR-10b-5p promote glioma cell migration and invasion both in vitro and in vivo.
a, b Analysis of glioma cell migration and invasion by transwell assay. a MiR-NC-, miR-1246-, and miR-10b-5p-transfected U87MG and U251 glioma cells that migrated to (without Matrigel coating) or invaded (with Matrigel coating) the bottom surface were stained with crystal violet and observed by light microscopy. Scale bar, 100 μm. b Quantitative analysis of miR-NC-, miR-1246-, and miR-10b-5p-transfected U87MG and U251 glioma cells that migrated to or invaded the bottom surface. c Analysis of glioma invasion by 3D invasion assay. Representative images of invaded spheroids in the 3D invasion assay for miR-NC-, miR-1246-, and miR-10b-5p-transfected U87MG and U251 glioma cells. Scale bar, 200 mm. d Quantitative analysis of area covered by miR-NC-, miR-1246-, and miR-10b-5p-transfected U87MG and U251 glioma cells. e Western blot analysis of N-cadherin, MMP2, MMP9, and vimentin in miR-NC-, miR-1246-, and miR-10b-5p-transfected U87MG and U251 glioma cells. f In vivo bioluminescent imaging of miR-NC-, miR-1246-, and miR-10b-5p-transfected U87MG glioma cells implanted in situ in nude mice. Representative images and quantification (25 days after implantation) are shown (five mice in each group). g HE staining of sections from mouse brain xenografts with miR-NC-, miR-1246-, and miR-10b-5p-transfected U87MG glioma cells. Scale bar, 200 μm. h Survival analysis for nude mice bearing miR-NC-, miR-1246-, and miR-10b-5p-transfected U87MG glioma cell xenografts (P < 0.05 by log-rank analysis). i Quantitative analysis of IHC staining for N-cadherin and MMP2 of sections from mouse brain xenografts with miR-NC-, miR-1246-, and miR-10b-5p-transfected U87MG glioma cells. j Quantitative analysis of IHC staining for Ki67 of sections from mouse brain xenografts with miR-NC-, miR-1246-, and miR-10b-5p-transfected U87MG glioma cells. Data are shown as the mean ± SD of three independent experiments. Statistical significance was determined using Student’s t test, ANOVA test and log-rank analysis (*P < 0.05; **P < 0.01).
MiR-1246 and miR-10b-5p enhance glioma migration and invasion by targeting FRK and TFAP2A
To explore the potential mechanisms by which miR-1246 and miR-10b-5p promote glioma migration and invasion, the online bioinformatic tools TargetScan and miRDB were used to predict the potential target genes of miR-1246 and miR-10b-5p. Among the predicted target genes, FRK and TFAP2A, which had been shown to be tumor suppressors and to be associated with tumor migration and invasion [29, 30], were chosen for further investigations. To determine whether FRK is a direct target of miR-1246 and TFAP2A is a direct target of miR-10b-5p, we confirmed the predicted binding sites in the 3′ untranslated region (UTR) of FRK and TFAP2A mRNA (Fig. 5a) and constructed the luciferase vectors containing the wild-type 3′UTR of FRK (FRK-WT), mutated 3′UTR of FRK (FRK-MUT), wild-type 3′UTR of TFAP2A (TFAP2A-WT), and mutated 3′UTR of TFAP2A (TFAP2A-MUT). The luciferase reporter assay results showed that miR-1246 could significantly attenuate FRK-WT luciferase activity, while it had a very modest effect on FRK-MUT in 293T cells (Fig. 5b), indicating FRK is a direct target of miR-1246. Similarly, miR-10b-5p could markedly reduce TFAP2A-WT luciferase activity but did not influence TFAP2A-MUT in 293T cells (Fig. 5b), suggesting that TFAP2A is a direct target of miR-10b-5p. Moreover, western blot analysis confirmed that miR-1246 inhibited FRK expression and miR-10b-5p suppressed TFAP2A expression in both U87MG and U251 glioma cells (Fig. 5c). To further confirm that miR-1246 and miR-10b-5p promote glioma migration and invasion by targeting FRK and TFAP2A, we knocked down FRK and TFAP2A by siRNAs in glioma cells. We found that both FRK and TFAP2A knockdown could significantly induce glioma migration and invasion (Fig. 5d, e). Furthermore, we restored FRK and TFAP2A expression by transfecting U87MG and U251 glioma cells with full-length FRK and TFAP2A plasmids. The migration assay showed that the effects of miR-1246 and miR-10b-5p on glioma migration were counteracted by FRK and TFAP2A overexpression (Fig. 5d, e). Western blot analysis also showed that miR-1246 and miR-10b-5p overexpression and FRK and TFAP2A knockdown could increase the expression of N-cadherin, MMP2, MMP9, and vimentin (Fig. 5f, g). Moreover, the effects of miR-1246 and miR-10b-5p could be attenuated by the overexpression of FRK and TFAP2A (Fig. 5f, g), indicating miR-1246 and miR-10b-5p induce glioma migration and invasion by directly targeting FRK and TFAP2A respectively (Fig. 6).
a Schematic representation of the predicted miR-1246 binding site in the 3’UTR of FRK, the mutated miR-1246 binding site in the 3′UTR of FRK, the predicted miR-10b-5p binding site in the 3′UTR of TFAP2A and the mutated miR-10b-5p binding site in the 3′UTR of TFAP2A. b Left: miR-NC or miR-1246 and luciferase vector encoding the wild-type or mutant 3′UTR of FRK were co-transfected into 293T cells and the relative luciferase activity was measured. Right: miR-NC or miR-10b-5p and luciferase vector encoding the wild-type or mutant 3′UTR of TFAP2A were co-transfected into 293T cells and the relative luciferase activity was measured. c Western blot analysis of FRK and TFAP2A in miR-NC-, miR-1246-, and miR-10b-5p-transfected U87MG and U251 glioma cells. d Representative images of indicated treated U87MG and U251 glioma cells that migrated (without Matrigel coating) to the bottom surface. Scale bar, 100 μm. e Quantification of indicated treated U87MG and U251 glioma cells that migrated (without Matrigel coating) to the bottom surface. f, g Western blot analysis of FRK, TFAP2A, N-cadherin, MMP2, MMP9, and vimentin levels in indicated treated U87MG and U251 glioma cells. Data are shown as the mean ± SD of three independent experiments. Statistical significance was determined using Student’s t test and one-way ANOVA test (*P < 0.05; **P < 0.01).
Hypoxia was an important feature during the progression of the glioma. Glioma cells in the hypoxic region released exosomes which contained miR-1246 and miR-10b-5p, which could be delivered to nomoxic glioma cells. The miR-1246 and miR-10b-5p promoted nomorxic glioma cell migration and invasion by directly targeting FRK and TFAP2A respectively.
Discussion
Exosomes are important in participate in intercellular communication. There are more and more studies focused on exosome functions in tumor progression and tumor microenvironment remodeling [31,32,33]. Interestingly, hypoxia, an important feature of the tumor microenvironment, has been proved to increase the production and alter the content of exosomes [18]; by this mechanism, hypoxia may modulate the functions and behavior of recipient cells. Many studies have demonstrated that hypoxic tumor exosomes could promote tumor growth [34], and facilitate tumor invasion and metastasis [35]. Furthermore, hypoxic tumor exosomes were reported to increase angiogenesis and vascular permeability [36], and suppress immune cells [20]. In the present study, we demonstrated that H-GDEs can promote normoxic glioma cell migration and invasion, which is consistent with results obtained with other tumors. Moreover, our results revealed the important roles of H-GDEs in glioma progression, suggesting the hypoxic glioma microenvironment induces normoxic glioma invasion and metastasis through glioma exosomes.
Being an important component of the tumor microenvironment, exosomes have been reported to participate in intercellular communication by delivering their content [12]. Many studies have demonstrated transfer of proteins and RNA molecules takes place between cells via exosomes [31, 35]. In this study, we confirmed miR-1246 and miR-10b-5p were increased significantly by hypoxia, exhibiting high expression levels in H-GDEs. In addition, our results indicate that miR-1246 and miR-10b-5p can be delivered to normoxic glioma tissue by H-GDEs. MiR-1246 and miR-10b-5p have been reported to exert inconsistent effects on tumor migration and invasion in several kinds of carcinomas and their function in glioma migration and invasion has not been well clarified [27, 37,38,39]. In the present study, we reveal for the first time that miR-1246 promotes glioma invasiveness. In addition, we have confirmed that miR-10b-5p enhances glioma invasive potential. These results indicate that miR-1246 and miR-10b-5p may be potential molecular targets for antitumor therapies. Given the important roles of miR-1246 and miR-10b-5p in promoting glioma migration and invasion, we also investigated whether miR-1246 and miR-10b-5p could serve as valuable biomarkers for the diagnosis and prognosis of glioma. We found both miR-1246 and miR-10b-5p expressions were positively associated with the glioma grade and glioma patients’ survival in CGGA database, suggesting miR-1246 and miR-10b-5p are potential biomarkers for glioma diagnosis and prognosis.
MicroRNAs exert their functions by directly targeting some mRNAs [40]. In our study, we have identified new targets of miR-1246 and miR-10b-5p in glioma. We demonstrated that miR-1246 and miR-10b-5p can promote glioma migration and invasion by targeting FRK and TFAP2A respectively. FRK has been reported to suppress glioma migration and invasion by regulating JNK/c-Jun signaling [29], and TFAP2A levels correlate inversely with glioma grade [30]. Consistent with these studies, our findings show that FRK and TFAP2A, as miR-1246 and miR-10b-5p targets, suppress glioma migration and invasion, suggesting that FRK and TFAP2A may act as suppressor molecules in many kinds of tumors.
In future studies, we will investigate the effects of other microRNAs in H-GDEs on glioma progression and their mechanisms of action. Furthermore, the development of a microfluidic chip for cerebrospinal fluid biopsy may improve glioma diagnosis by including microRNAs such as miR-1246 and miR-10b-5p. In conclusion, we have demonstrated that H-GDE-derived miR-1246 and miR-10b-5p can promote glioma migration and invasion by targeting FRK and TFAP2A respectively. Our findings also disclose miR-1246 and miR-10b-5p may be biomarkers for glioma diagnosis and prognosis. Treatment targeting miR-1246 and miR-10b-5p may impair the motility of glioma cells, representing a novel direction for the development of antitumor therapy.
References
Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011-2015. Neuro Oncol. 2018;20:iv1–iv86.
Prados MD, Byron SA, Tran NL, Phillips JJ, Molinaro AM, Ligon KL, et al. Toward precision medicine in glioblastoma: the promise and the challenges. Neuro Oncol. 2015;17:1051–63.
Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia. 2012;60:502–14.
Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99:1441–54.
Lu X, Kang Y. Hypoxia and hypoxia-inducible factors: master regulators of metastasis. Clin Cancer Res. 2010;16:5928–35.
Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Investig. 2010;120:127–41.
Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95.
Harris AL. Hypoxia-a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47.
Pennacchietti S, Michieli P, Galluzzo M, Mazzone M, Giordano S, Comoglio PM. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 2003;3:347–61.
Chang Q, Jurisica I, Do T, Hedley DW. Hypoxia predicts aggressive growth and spontaneous metastasis formation from orthotopically grown primary xenografts of human pancreatic cancer. Cancer Res. 2011;71:3110–20.
Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–79.
Tkach M, Thery C. Communication by Extracellular Vesicles: where We Are and Where We Need to Go. Cell. 2016;164:1226–32.
Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–30.
Casadei L, Calore F, Creighton CJ, Guescini M, Batte K, Iwenofu OH, et al. Exosome-Derived miR-25-3p and miR-92a-3p Stimulate Liposarcoma Progression. Cancer Res. 2017;77:3846–56.
Zhang HG, Grizzle WE. Exosomes and cancer: a newly described pathway of immune suppression. Clin Cancer Res. 2011;17:959–64.
Graner MW, Schnell S, Olin MR. Tumor-derived exosomes, microRNAs, and cancer immune suppression. Semin Immunopathol. 2018;40:505–15.
Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–801.
King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421.
Li L, Li C, Wang S, Wang Z, Jiang J, Wang W, et al. Exosomes Derived from Hypoxic Oral Squamous Cell Carcinoma Cells Deliver miR-21 to Normoxic Cells to Elicit a Prometastatic Phenotype. Cancer Res. 2016;76:1770–80.
Guo X, Qiu W, Liu Q, Qian M, Wang S, Zhang Z, et al. Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten Pathways. Oncogene. 2018;37:4239–59.
Bang C, Batkai S, Dangwal S, Gupta SK, Foinquinos A, Holzmann A, et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Investig. 2014;124:2136–46.
Xue H, Yuan G, Guo X, Liu Q, Zhang J, Gao X, et al. A novel tumor-promoting mechanism of IL6 and the therapeutic efficacy of tocilizumab: Hypoxia-induced IL6 is a potent autophagy initiator in glioblastoma via the p-STAT3-MIR155-3p-CREBRF pathway. Autophagy. 2016;12:1129–52.
Roy R, Yang J, Moses MA. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J Clin Oncol. 2009;27:5287–97.
Song H, Li Y, Lee J, Schwartz AL, Bu G. Low-density lipoprotein receptor-related protein 1 promotes cancer cell migration and invasion by inducing the expression of matrix metalloproteinases 2 and 9. Cancer Res. 2009;69:879–86.
Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518–29.
Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6.
Li D, Zhang Y, Zhang H, Zhan C, Li X, Ba T, et al. CADM2, as a new target of miR-10b, promotes tumor metastasis through FAK/AKT pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:46.
Yang F, Xiong H, Duan L, Li Q, Li X, Zhou Y. MiR-1246 Promotes Metastasis and Invasion of A549 cells by Targeting GSK-3β‒Mediated Wnt/β-Catenin Pathway. Cancer Res Treat. 2019;51:1420–29.
Zhou X, Hua L, Zhang W, Zhu M, Shi Q, Li F, et al. FRK controls migration and invasion of human glioma cells by regulating JNK/c-Jun signaling. J Neurooncol. 2012;110:9–19.
Heimberger AB, McGary EC, Suki D, Ruiz M, Wang H, Fuller GN, et al. Loss of the AP-2alpha transcription factor is associated with the grade of human gliomas. Clin Cancer Res. 2005;11:267–72.
Fang T, Lv H, Lv G, Li T, Wang C, Han Q, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191.
Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin YF, Yuan Y, et al. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. 2018;68:1459–75.
Fu Q, Zhang Q, Lou Y, Yang J, Nie G, Chen Q, et al. Primary tumor-derived exosomes facilitate metastasis by regulating adhesion of circulating tumor cells via SMAD3 in liver cancer. Oncogene. 2018;37:6105–18.
Xue M, Chen W, Xiang A, Wang R, Chen H, Pan J, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16:143.
Shan Y, You B, Shi S, Shi W, Zhang Z, Zhang Q, et al. Hypoxia-Induced Matrix Metalloproteinase-13 Expression in Exosomes from Nasopharyngeal Carcinoma Enhances Metastases. Cell Death Dis. 2018;9:382.
Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC, Tsai PH, et al. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36:4929–42.
Bhagirath D, Yang TL, Bucay N, Sekhon K, Majid S, Shahryari V, et al. microRNA-1246 Is an Exosomal Biomarker for Aggressive Prostate Cancer. Cancer Res. 2018;78:1833–44.
Lin SS, Peng CY, Liao YW, Chou MY, Hsieh PL, Yu CC. miR-1246 Targets CCNG2 to Enhance Cancer Stemness and Chemoresistance in Oral Carcinomas. Cancers (Basel). 2018;10:272.
He C, Chen ZY, Li Y, Yang ZQ, Zeng F, Cui Y, et al. miR-10b suppresses cell invasion and metastasis through targeting HOXA3 regulated by FAK/YAP signaling pathway in clear-cell renal cell carcinoma. BMC Nephrol. 2019;20:127.
Chen X, Zhou J, Li X, Wang X, Lin Y, Wang X. Exosomes derived from hypoxic epithelial ovarian cancer cells deliver microRNAs to macrophages and elicit a tumor-promoted phenotype. Cancer Lett. 2018;435:80–91.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81874083; 82072776; 82072775; 81702468; 81802966; 81902540), Natural Science Foundation of Shandong Province of China (No. ZR2019BH057), Key clinical Research project of Clinical Research Center of Shandong University (2020SDUCRCA011) and Taishan Scholars of Shandong Province of China (No. ts201511093).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Qian, M., Chen, Z., Guo, X. et al. Exosomes derived from hypoxic glioma deliver miR-1246 and miR-10b-5p to normoxic glioma cells to promote migration and invasion. Lab Invest 101, 612–624 (2021). https://doi.org/10.1038/s41374-020-00522-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-020-00522-0
This article is cited by
-
Extracellular Vesicles Derived from Glioma Stem Cells Affect Glycometabolic Reprogramming of Glioma Cells Through the miR-10b-5p/PTEN/PI3K/Akt Pathway
Stem Cell Reviews and Reports (2024)
-
Crucial role of the transcription factors family activator protein 2 in cancer: current clue and views
Journal of Translational Medicine (2023)
-
Spheresomes are the main extracellular vesicles in low-grade gliomas
Scientific Reports (2023)
-
Tumor-derived small extracellular vesicles: potential roles and mechanism in glioma
Journal of Nanobiotechnology (2022)
-
The hypoxia-driven crosstalk between tumor and tumor-associated macrophages: mechanisms and clinical treatment strategies
Molecular Cancer (2022)