Abstract
Cardiovascular diseases are considered the leading cause of death worldwide. Myocardial ischaemia/reperfusion (I/R) injury is recognized as a critical risk factor for cardiovascular diseases. Although increasing advances have been made recently in understanding the mechanisms of I/R injury, they remain largely unknown. In this study, we found that the expression of circPAN3 (circular RNA PAN3) was decreased in a mouse model of myocardial I/R. Overexpression of circPAN3 significantly inhibited autophagy and alleviated cell apoptosis of cardiomyocytes, which was further verified in vivo by decreased autophagic vacuoles and reduced myocardial infarct sizes. Moreover, miR-421 (microRNA-421) was identified as a downstream target involved in circPAN3-mediated myocardial I/R injury. Additionally, miR-421 could negatively regulate Pink1 (phosphatase and tensin homologue-induced putative kinase 1) via a direct binding relationship. Furthermore, the mitigating effects of circPAN3 overexpression on myocardial I/R injury by suppressing autophagy and apoptosis were abolished by knockdown of Pink1. Our findings reveal a novel role for circPAN3 in modulating autophagy and apoptosis in myocardial I/R injury and the circPAN3–miR-421–Pink1 axis as a regulatory network, which might provide potential therapeutic targets for cardiovascular diseases.
Similar content being viewed by others
Introduction
Cardiovascular diseases (CVDs), such as myocardial infarction, stroke, rheumatic heart disease and coronary heart disease, affect millions of patients and are considered the leading cause of death worldwide [1]. Ischaemia/reperfusion (I/R) injury is the tissue damage caused by the initial ischaemia and subsequent injury as a result of reperfusion, which is closely associated with several CVDs, including myocardial infarction, leading to increased morbidity and mortality [2, 3]. Myocardial ischaemia is caused by the obstruction of blood flow to the heart, generally due to coronary thrombosis, stenosis or vasospasm, resulting in the imbalance of myocardial oxygen demand and supply [4]. Myocardial ischaemia can give rise to serious and fatal complications, including myocardial infarction, cardiac failure and arrhythmia. Myocardial ischaemia can be treated with medications, such as aspirin and nitrates, and surgery including angioplasty and stenting to improve blood flow to the heart [5]. However, these treatments might cause I/R injury to patients. Although increasing advances have been made recently in understanding the mechanisms of I/R injury, such as oxidative stress and the inflammatory response [6, 7], they still remain largely unknown. Therefore, elucidating the pathological mechanisms for the occurrence and development of I/R injury is extremely important for developing novel and effective therapeutic strategies to control I/R injury.
Autophagy, a highly conserved self-digesting cellular process, refers to lysosomal degradation of intracellular components such as damaged organelles and pathogenic proteins in response to various stresses and has been categorized into three types: microautophagy, macroautophagy and chaperone-mediated autophagy [8]. Autophagic cell death differs from apoptosis and necrosis [9]. Autophagy plays important roles in regulating multiple physiological processes, including cell growth, differentiation, death and survival, and maintaining cellular homoeostasis [8, 10], and its dysregulation has been implicated in diseases such as cancers and neurodegenerative and infectious diseases [11, 12]. It has also been demonstrated that autophagy is associated with various CVDs [13]. For instance, Valentim et al. [14] reported that suppression of beclin expression, a pro-autophagic gene, reduced I/R-induced autophagy and enhanced cardiomyocyte survival. Although autophagy has been well recognized to play a crucial role in CVDs, there are still no very effective therapies targeting autophagy in CVD treatment. Therefore, a better understanding of the mechanisms of autophagy in CVDs might yield novel cellular therapeutic targets.
Unlike the better known linear RNAs, circular RNA (circRNA), a single-stranded non-coding RNA that is abundant and evolutionarily conserved, forms a closed circle by the covalent linkage between its 5′ and 3′ ends and is produced via back-splicing circularization across introns, exons or both [15]. Growing evidence indicates that circRNAs act as a microRNA (miRNA) sponge that sequester miRNAs to regulate gene expression and play important roles in human diseases [16,17,18]. Although it has become a research focus since 2012, the expression patterns and molecular mechanisms of circRNAs in I/R injury are still largely unknown. Our study focused on circPAN3, originating from the Pan3 transcript, which regulates the self-renewal of intestinal stem cells [19] and facilitates the drug resistance of acute myeloid leukaemia by modulating autophagy [20]. miR-421 is a well-studied miRNA, and it has been reported that miR-421 is associated with heart function [21] and human cancers [22, 23]. Moreover, Wang et al. demonstrated that E2F1 (E2F transcription factor 1)-activated miR-421 promoted mitochondrial fragmentation, apoptosis and myocardial infarction by suppressing the translation of Pink1 [24], a regulator of autophagy in cardiomyocytes [25], indicating the role of miR-421 in CVDs. However, whether circPAN3 is involved in CVD pathogenesis and its potential mechanisms remain elusive.
In this study, we demonstrated for the first time that circPAN3 ameliorates myocardial I/R injury by absorbing miR-421 as a competing endogenous RNA (ceRNA) and regulating Pink1-mediated autophagy. Our investigation not only revealed the vital roles of the circPAN3/miR-421/Pink1 axis in I/R injury but also provided potential diagnostic markers of I/R injury and promising therapeutic targets for patients with I/R injury.
Methods
Mouse model of myocardial I/R injury
The mouse model of myocardial I/R injury was established as previously described [24, 26]. Briefly, 100 male C57BJ/6L mice at the age of 10–12 weeks (purchased from Charles River, Beijing, China) were first fed a mouse diet and given tap water in a temperature-controlled room (12/12 h light/dark cycle). Then, mice (n = 10 per group) were randomly separated into five groups: (1) a sham group; (2) an I/R group; (3) an I/R + vector group; (4) an I/R + circPAN3 group; and 5) an I/R + circPAN3 + siPink1 group. Before the thoracotomy, mice were anaesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg). The heart was exposed, and the left anterior descending artery (LAD) was located with a dissection microscope. Then, a 6-0 silk suture was passed around the LAD, and a loose knot was made at the inferior border of the left auricle. A PE10 tubing was placed between the knot and the vessel. The LAD was occluded by tightening the loop for 45 min of ischaemia. After ischaemia, the loop was untied, and the PE10 tubing was removed for 3 h of reperfusion. The knot was left in place for subsequent triphenyl tetrazolium chloride (TTC) staining. The procedure for the sham group was the same except that the knot was untied. Hearts were excised from mice and processed for subsequent assays. All animal procedures were approved by the Institutional Animal Care and Use Committee of Xiangya Hospital of Central South University and conducted in accordance with the National Institutes of Health guidelines.
For gene modification in vivo, circPAN3 and Pink1 cDNA were cloned into the Adeno-X expression system (Clontech, Mountain View, CA, USA). The adenoviruses harbouring siRNAs against Pink1 (siPink1) were constructed using the pSilencer adeno 1.0-CMV System (Ambion, Grand Island, NY, USA). Mice were administered 5.0 × 1010 plaque-forming units (pfu) of circPAN3, circPAN3/Pink1 or circPAN3/siRNA adenoviruses in the tail vein. Five days after adenovirus administration, mice were subjected to I/R treatment.
Cell culture, treatment and transfection
Primary human cardiac myocytes (HCMs) and embryonic kidney 293 (HEK293) cells were purchased from PromoCell (Heidelberg, Germany) and the American Type Culture Collection (ATCC, Manassas, VA, USA), respectively. Cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% foetal bovine serum at 37 °C in a cell incubator with 5% CO2. All of the above cell culture reagents were purchased from Thermo Fisher Scientific (Waltham, MA, USA). For hypoxia/reoxygenation (H/R) treatment, HCMs were cultured in an anaerobic chamber containing 95% N2 and 5% CO2 for 0, 8, 16 or 24 h and then reoxygenated for 12 h.
For cell transfection, HCMs were seeded in six-well plates, cultured to approximately 80% confluency and transfected with the circPAN3 overexpression vector (circPAN3), empty vector, miR-421 mimics, mimics control (miR-NC), miR-421 inhibitor (anti-miR-421) or inhibitor control (anti-miR-NC). Then, the cells were harvested for subsequent analysis. miR-421 mimics, mimics control, miR-421 inhibitor and inhibitor control were ordered from GenePharma (Shanghai, China).
Enzyme activity measurement
For the measurement of enzyme activity, including lactic dehydrogenase (LDH), creatine kinase(CK) and creatine kinase MB isoenzyme (CK-MB), serum samples were prepared from mice in the I/R injury or sham group, and 10 µL of serum was used for the measurement. The measurement was performed following the manufacturer’s instructions. For CK-MB activity detection, an anti-M subunit polyclonal antibody was used to block the M subunits of CK-MB and CK-MM (creatine kinase MM isoenzyme). The absorbance at 450 nm for LDH and 340 nm for CK and CK-MB was measured. Kits were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Echocardiographic assessment
After 1 week of the I/R procedure or sham operation, mice were subjected to transthoracic echocardiography as previously described [27]. The following echocardiographic parameters were recorded: left ventricular end-diastolic dimension (LVEDD), end-systolic diameters (LVESD), end‐diastolic pressure (LVEDP, which was calculated by subtracting the gradient across the aortic valve at the end of diastole from the diastolic blood pressure with the online calculator: https://e-echocardiography.com/page/page.php?UID=175716901), ejection fraction (LVEF, which was calculated by subtracting the LV volume in systole from the LV volume in diastole and then dividing the difference by the LV volume in diastole), systolic pressure (LVSP, which was estimated from mitral valve regurgitation velocity using the simplified Bernoulli equation), systolic internal diameter (LVIDs) and diastolic internal diameters (LVIDd). Fractional shortening of left ventricular diameter (LVFS) was calculated with the following formula: LVFS = [(LVIDd/LVIDs)/LVIDd] × 100%.
Histological staining
Hearts were harvested from mice with I/R injury and the sham group, fixed in 3.7% (v/v) formaldehyde, dehydrated and embedded in paraffin and were then cut into five-micron tissue sections. For haematoxylin and eosin (H&E) staining, sections were deparaffinized and stained with haematoxylin and eosin. Images were captured with an Olympus BX51 microscope (Tokyo, Japan). For terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL), the TUNEL Colorimetric IHC Detection Kit was ordered from Thermo Fisher Scientific (Waltham, MA, USA). The TUNEL assay was performed following the manufacturer’s recommendation. In brief, sections were deparaffinized, fixed, permeabilized and subjected to TdT and Click-iT™ reactions. Then, sections were washed, dried and mounted. Sections were imaged with an Olympus BX51 microscope.
TTC staining
After the sham procedure or I/R surgery, hearts were exposed, rapidly excised, frozen and cut into slices of 1 mm. Then, the slices were incubated with 2% TTC solution for 40 min at 37 °C. The stained slices were fixed again in 10% formaldehyde overnight and imaged. TTC-stained and non-TTC-stained areas were analysed with Image-Pro Plus image analysis 6.0 software. The ratio of infarcted myocardium to the risk region was calculated as the whole myocardial tissues (infarct area/whole heart area) × percentage.
Transmission electron microscopy (TEM)
Hearts were excised from mice with I/R surgery or sham operation and immediately fixed in 3% glutaraldehyde overnight. Samples were then washed, post-fixed, dehydrated, embedded in EMbed 812 resin (VWR, Radnor, PA) and cut into 80-nm-thick sections, which were placed on 200 mesh copper grids for TEM (Sigma-Aldrich). Uranyl acetate and lead citrate were used to stain the grids. Samples were viewed with a Krios transmission electron microscope (FEI, Hillsboro, OR, USA).
Immunofluorescence assay
HCMs were plated on chamber slides, treated with H/R and transfected as indicated. Then, myocytes were washed, fixed in 4% formaldehyde for 15 min at room temperature and permeabilized with 0.2% Triton X-100 for 10 min at room temperature. After permeabilization, cells were washed, blocked and incubated with anti-human LC3 (light chain 3) antibody (Cell Signaling Technology, Boston, MA, USA) at 4 °C overnight. The next day, the cells were probed with an Alexa Fluor 488-conjugated goat anti-rabbit IgG (immunoglobulin G) secondary antibody, stained with DAPI (4′,6-diamidino-2-phenylindole) and mounted. Slides were imaged with a Nikon confocal microscope.
TUNEL assay
The TUNEL assay was performed following the manufacturer’s instructions. HCMs were plated on chamber slides, treated with H/R and transfected as indicated. Cells were washed, fixed in 4% paraformaldehyde for 15 min and permeabilized in 0.25% Triton X-100 for 20 min at room temperature. Then, cells were subjected to the TdT (terminal deoxynucleotidyl transferase) reaction followed by the Click-iT reaction, stained with DAPI and mounted. Slides were imaged with a Nikon confocal microscope.
Caspase 3 activity analysis
The caspase 3 activity colorimetric kit was purchased from Clontech Laboratories (Palo Alto, CA, USA). Caspase 3 activity analysis was performed according to the manufacturer’s protocol. Briefly, HCMs with the indicated treatment and transfection were collected and lysed with lysis buffer. Fifty microlitres of cell lysates (a total of 20 µg protein) was transferred into a new well. Then, 50 µL of reaction buffer and 5 µL of caspase 3 substrate were added. Samples were incubated at 37 °C for an hour. The absorbance was read at 405 nm with a microplate reader.
Vector construction
The circPAN3 sequence along with the 1 kb upstream flanking sequence was synthesized by Sangon Biotech (Shanghai, China) and inserted into pcDNA3.1. Then, the upstream flanking sequence was inserted in an inverted orientation downstream. Adenoviruses harbouring circPAN3 or Pink1 siRNA constructs were generated with the Adeno-X™ Expression System (Clontech Laboratories) or pSilencer adeno 1.0-CMV System (Thermo Fisher Scientific), respectively, following the manufacturer’s instructions.
Real-time quantitative reverse transcription-PCR (RT-qPCR)
Total RNA was extracted from mouse heart tissues and HCMs with TRIzol reagent (Thermo Fisher Scientific) and reverse-transcribed into cDNA. miR-421 was reverse transcribed using a TaqMan microRNA reverse transcription kit (Thermo Fisher Scientific). The relative expression levels of circPAN3, miR-421 and Pink1 were analysed with SYBR Green QPCR Master Mix (Toyobo, Osaka, Japan) using a 7500 Real Time PCR System (Thermo Fisher Scientific). The results were normalized to GAPDH (glyceraldehyde 3 phosphate dehydrogenase) or U6 snRNA using the 2−ΔΔCt method. The primers for miR-421, circPAN3 and U6 snRNA were purchased from RiboBio (Guangzhou, China). Other primers are listed in Table 1.
Western blotting
Total protein was extracted from HCMs or heart tissues using radioimmunoprecipitation assay lysis buffer (Abcam) and quantified with a BCA protein quantitation kit (Thermo Fisher Scientific). Forty micrograms of cell lysates were loaded, electrophoresed and transferred to polyvinylidene fluoride membranes (GE Healthcare, Pittsburgh, PA, USA). Membranes were blocked and probed with primary antibodies against LC3 (human, 1:1000; mouse 1:800), Beclin1 (Bcl-2-interacting myosin-like coiled-coil protein, human and mouse, 1:1000), P62 (human and mouse, 1:500), Bcl-2 (B-cell lymphoma 2, human, 1:500; mouse, 1:1000), Bax (BCL2-associated X protein, human, 1:1000; mouse, 1:500), pro-caspase 3 (human and mouse, 1:1000), cleaved caspase 3 (human and mouse, 1:1000) and GAPDH (human and mouse, 1:5000) for 2 h at room temperature. All primary antibodies were ordered from Cell Signaling Technology (Boston, MA, USA). Blots were then rinsed and incubated with HRP (horseradish peroxidase)-conjugated secondary antibodies (Abcam) and visualized with enhanced chemiluminescence western blotting substrate (Thermo Fisher Scientific). The relative band intensity was analysed with ImageJ software (NIH).
Dual-luciferase reporter assay
The predicted binding sites of miR-421 in circPAN3 (circPAN3-WT) and Pink1 (Pink1-WT) and the corresponding mutated sequences (circPAN3-MUT and Pink1-MUT) were separately cloned into the pmirGLO vector (Promega, Madison, WI, USA). HEK293 cells were co-transfected with the above constructs and miR-421 mimics or miR-421 NC and harvested. Then, the firefly and Renilla luciferase activities were examined with the Dual-Glo® Luciferase Assay System (Promega) following the manufacturer’s instructions. The relative firefly luciferase activity was normalized to Renilla luciferase activity.
RNA pull-down assay
RNA pull-down was performed as previously described [28]. The biotinylated circPAN3 probe was specifically designed to bind to the junction area of circPAN3, and the oligo probe was used as the negative control. The circPAN3 probe (Tsingke, Wuhan, China) was incubated with streptavidin magnetic beads (Life Technologies, USA) at room temperature for 2 h to generate probe-coated beads, which were then added to the cell lysates at 4 °C overnight. Then, the beads were washed, and the bound miRNA was extracted using TRIzol reagent and analysed by qRT-PCR.
For the biotinylated miR-421 probe, briefly, HCMs were transfected with biotinylated miR-421 mimics or mimics control and harvested for cell lysis on ice for 30 min. The cell lysates were incubated with magnetic streptavidin beads for 20 h at 4 °C. Then, the bound RNA was recovered and analysed by RT-PCR. GAPDH was used as a negative control. The biotinylated miR-421 mimics or mimics control were synthesized by Sangon Biotech.
Statistical analysis
All data in this study were analysed and graphed using Prism 6.0 software (GraphPad Software, San Diego, CA, USA) and expressed as the mean ± standard deviation of at least three independent experiments. Student’s t-test for comparing two independent groups and one-way analysis of variance (ANOVA) for multiple group comparisons were performed for statistical analyses. A value of P < 0.05 was considered statistically significant.
Results
Enhanced autophagy and downregulation of circPAN3 were observed in a mouse model of myocardial I/R injury
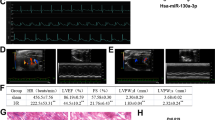
To investigate the roles of circPAN3 in myocardial I/R injury, a mouse model of myocardial I/R injury was successfully established as previously described [24, 26]. Compared with the sham group, mice with I/R injury showed increased bleeding and neutrophil infiltration, disordered arrangement of myocardial bundles (Fig. 1a) and enhanced myocardial infarct (Fig. 1b). In addition, we found increased autophagic vacuoles (Fig. 1c) and an enhanced ratio of LC3II/LC3I expression (Fig. 1d), indicating that autophagy was significantly promoted in mice with I/R treatment. The enhanced activity of LDH, CK and CK-MB (Fig. 1e) and abnormal echocardiographic parameters, including LVEDD, LVESD, LVEDP, LVEF, LVSP and LVFS (Fig. 1f), implied that heart function was severely impaired upon I/R injury. Intriguingly, the expression of circPAN3 was significantly downregulated in mice with I/R injury compared with the sham group (Fig. 1g). These results raised the possibility that circPAN3 and autophagy might be associated with myocardial I/R injury.
a H&E staining of heart sections from I/R-treated mice and the sham group. b TTC staining of heart sections from I/R-treated mice and the sham group (n = 10). c The examination of autophagic vacuoles by TEM in heart tissues from I/R-treated mice compared with the sham group (n = 10). d Western blot analysis was performed to assess the level of LC3II/I. e Cardiac enzyme measurement in serum from I/R-treated mice compared with the sham group (n = 10). f Echocardiographic assessment of heart function in I/R-treated mice compared with the sham group (n = 10). g Quantitative RT-PCR analysis of circPAN3 (n = 10). GAPDH was used as a normalization control in western blot analysis and quantitative RT-PCR analysis. All data were from at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
Overexpression of circPAN3 ameliorated myocardial I/R injury in vivo
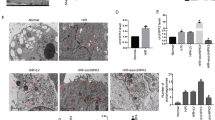
To further study the relationship of circPAN3, autophagy and myocardial I/R injury, circPAN3 was overexpressed in mice with I/R treatment. Compared with the vector control, the overexpression of circPAN3 markedly alleviated the structural damage of myocardial tissue and reduced the myocardial infarct size (Fig. 2a, b) caused by I/R treatment. Moreover, the number of autophagic vacuoles, the area occupied by autophagic vacuoles and the expression of LC3II were decreased upon circPAN3 overexpression (Fig. 2c, d), indicating that circPAN3 might regulate autophagy in myocardial I/R injury. The enhanced activity of LDH, CK and CK-MB caused by I/R treatment was inhibited (Fig. 2e), and heart function assessed by echocardiography was also restored by circPAN3 overexpression (Fig. 2f). The overexpression of circPAN3 in mice with I/R injury was validated using the RT-qPCR assay (Fig. 2g). These results demonstrated that circPAN3 might regulate autophagy to exert positive effects on myocardial I/R injury.
a H&E staining of heart sections from sham, I/R-treated mice (I/R), I/R-treated mice with empty vector transfection (I/R + vector) and I/R-treated mice with circPAN3 expression vector transfection (I/R + circPAN3). b TTC staining of heart sections from the indicated mice (n = 10). c The examination of autophagic vacuoles by TEM in heart tissues from the indicated mice (n = 10). d Western blot analysis of LC3II/I in the indicated mice. e Cardiac enzyme measurement in serum from the indicated mice (n = 10). f Echocardiographic assessment of heart function in the indicated mice (n = 10). g Quantitative RT-PCR analysis of circPAN3 in the indicated mice (n = 10). GAPDH was used as a normalization control in western blot analysis and quantitative RT-PCR analysis. All data were from at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
Overexpression of circPAN3 alleviated H/R-induced cardiomyocyte damage by suppressing autophagy and apoptosis in vitro
As I/R injury is mainly due to hypoxia and reoxygenation [29], HCMs were treated with H/R as the in vitro cell model of myocardial I/R injury. The expression of circPAN3 was analysed using RT-qPCR. We found that along with the increase in H/R treatment time (0, 8, 16 and 24 h), the expression of circPAN3 was significantly inhibited consistent with its expression pattern in mice with I/R injury (Fig. 3a). As expected, H/R treatment increased the expression of LC3II and its recruitment to autophagosomal membranes, whereas this induction was abolished by overexpression of circPAN3 (Fig. 3b). As autophagy is closely associated with cell apoptosis [30], we analysed the expression of LC3, Beclin1, P62, Bcl-2, Bax and Caspase 3. Compared with controls, H/R treatment promoted the expression of autophagy markers (LC3II and Beclin1) and pro-apoptotic factors (Bax and cleaved caspase 3) and suppressed the expression of P62 and the anti-apoptotic factor Bcl-2, which were reversed by the overexpression of circPAN3 (Fig. 3c). The TUNEL assay and the measurement of caspase 3 activity also showed increased apoptosis after H/R treatment, and apoptosis was suppressed by circPAN3 overexpression (Fig. 3d, e). These observations demonstrated that circPAN3 might alleviate cardiomyocyte damage by inhibiting autophagy and apoptosis in myocardial I/R injury.
a Quantitative RT-PCR analysis of circPAN3 in HCMs treated with H/R for the indicated times (n = 3). b Immunofluorescence staining of LC3I/II in HCMs in response to control treatment (Con), H/R treatment (H/R), H/R treatment plus empty vector transfection (H/R + vector) and H/R treatment plus circPAN3 expression vector transfection (H/R + circPAN3). c Western blot analysis of LC3I/II, Beclin1, P62, Bax, Bcl-2 and cleaved caspase 3 in HCMs with the indicated treatment. d TUNEL staining of HCMs with the indicated treatments (n = 3). e The assessment of caspase 3 activity in HCMs with the indicated treatments (n = 3). GAPDH was used as a normalization control in western blot analysis and quantitative RT-PCR analysis. All data were from at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
CircPAN3 acts as a sponge of miR-421
Since circPAN3 generally acts as a ceRNA to absorb miRNAs, we predicted its downstream target by bioinformatics analysis and identified a highly likely binding site for miR-421 (Fig. 4a). The luciferase reporter assay showed that luciferase activity was markedly reduced upon cotransfection with miR-421 mimics and the wild-type circPAN3 construct but unaffected by the mutated circPAN3 construct (Fig. 4b), implying that miR-421 is a functional direct target of circPAN3. We found that the expression of miR-421 was significantly decreased in the circPAN3-overexpressing HCMs (Fig. 4c). Subsequently, we performed a biotin-labelled probe pull-down assay to investigate whether circPAN3 could directly bind miR-421. First, the biotin-labelled probe was verified to pull down circPAN3 in HCMs (Fig. 4d). Next, biotin-labelled miR-421 was used to confirm its direct binding relationship, and the results showed that biotin-labelled miR-421 captured more circPAN3 than the negative control (Fig. 4e). We also analysed miR-421 expression in mice and found that miR-421 was upregulated upon I/R treatment compared with sham treatment (Fig. 4f). In addition, the expression of circPAN3 and miR-421 was negatively correlated in mice with I/R injury (Fig. 4g). These data demonstrated that circPAN3 might directly target miR-421 to exert its roles in myocardial I/R injury.
a The predicted binding site of circPAN3 and miR-421. b Analysis of the relative luciferase activity (n = 3). HEK293 cells were transfected with circPAN3-WT/miR-421, circPAN3-WT/miR-NC, circPAN3-MUT/miR-421 or circPAN3-MUT/miR-NC. c Quantitative RT-PCR analysis of miR-421 in HCMs transfected with vector only or the circPAN3 vector (n = 3). U6 RNA was used as a normalization control. d CircPAN3 in HCM lysates was pulled down and enriched with a circPAN3-specific probe and then examined by semi-quantitative RT-PCR assay. GAPDH was used as a control. e Biotin-labelled miR-421 was transfected into HCMs, and circPAN3 levels were quantified by quantitative RT-PCR (n = 3). f Quantitative RT-PCR analysis of miR-421 in I/R-treated mice and sham mice (n = 10). GAPDH was used as a normalization control. g Pearson correlation analysis of circPAN3 and miR-421. All data were from at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
miR-421 reversed circPAN3-mediated protective effects on H/R-induced cardiomyocyte damage
To assess whether miR-421 is the mediator of circPAN3 exerting its effects in I/R injury, H/R-treated HCMs were transfected with miR-421 mimics alone or co-transfected with miR-421 mimics and the circPAN3 expression vector. Overexpression of circPAN3 dramatically impaired the activation of autophagy induced by H/R treatment, while this inhibitory effect was reversed by miR-421 mimics (Fig. 5a). Consistent with autophagy, apoptosis was also increased by upregulation of miR-421, but this effect was reversed by circPAN3 transfection (Fig. 5b, c), suggesting that circPAN3 might regulate autophagy and apoptosis by targeting miR-421 in I/R injury. Indeed, the expression of miR-421 was increased in HCMs with H/R treatment alone and further enhanced by miR-421 overexpression but inhibited by circPAN3 overexpression compared with that in the H/R treatment group (Fig. 5d). Similarly, the effect of H/R treatment on the protein expression of LC3, Beclin1, P62, Bcl-2, Bax and Caspase 3 was strengthened by miR-421 overexpression and decreased by circPAN3 transfection (Fig. 5e). These results demonstrated that circPAN3 directly absorbed miR-421 to regulate autophagy in myocardial I/R injury.
a Immunofluorescence staining of LC3 in HCMs in response to Con, H/R, H/R + circPAN3, H/R treatment plus miR-421 mimic transfection (H/R + miR-421) and H/R + circPAN3+miR-421. b TUNEL staining of HCMs with the indicated treatments (n = 3). c The assessment of caspase 3 activity in HCMs with the indicated treatment (n = 3). d Quantitative RT-PCR analysis of miR-421 in HCMs with the indicated treatments (n = 3). U6 RNA was used as a normalization control. e Western blot analysis of LC3II/I, Beclin1, P62, Bax, Bcl-2 and caspase 3 in HCMs with the indicated treatments. GAPDH was used as a normalization control. All data were from at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
Pink1 was a target of miR-421
Along with the extension of H/R treatment time (0, 8, 16 and 24 h), the expression of miR-421 was significantly increased (Fig. 6a). As it has been reported that miR-421 facilitates cardiomyocyte apoptosis and myocardial infarction by suppressing the expression of Pink1 [24], we analysed Pink1 expression and found that the mRNA and protein levels of Pink1 were inhibited by H/R treatment and that this effect was largely reversed by miR-421 inhibitor transfection (Fig. 6b, c). The potential binding of miR-421 and Pink1 was predicted by bioinformatics analysis (Fig. 6d). Luciferase activity was obviously suppressed upon cotransfection with miR-421 mimics and the wild-type 5′UTR of Pink1 but was unaffected by the mutated Pink1 construct (Fig. 6e), indicating that Pink1 was the functional direct target of miR-421 (Fig. 6e). The expression of Pink1 was reduced by miR-421 mimics but promoted by the miR-421 inhibitor (Fig. 6f). In addition, Pink1 was upregulated upon I/R treatment (Fig. 6g), and the expression of Pink1 and miR-421 was negatively correlated in mice with I/R injury (Fig. 6h). These data demonstrated that miR-421 directly bound Pink1 to negatively regulate its expression in myocardial I/R injury.
a Quantitative RT-PCR analysis of miR-421 in HCMs with H/R treatment for the indicated times (n = 3). U6 RNA was used as a normalization control. b Quantitative RT-PCR analysis of Pink1 in HCMs in response to Con, H/R, H/R plus anti-miR-421 NC transfection (H/R + NC) and H/R plus anti-miR-421 transfection (H/R + anti-miR-421) (n = 3). U6 RNA was used as a normalization control. c Western blot analysis of Pink1 in HCMs with the indicated treatment. GAPDH was used as a normalization control. d The predicted binding site of miR-421 in the 5′UTR of Pink1. e Analysis of the relative luciferase activity (n = 3). HEK293 cells were transfected with Pink1-WT/miR-421, Pink1-WT/miR-NC, Pink1-MUT/miR-421 or Pink1-MUT/miR-NC. f Quantitative RT-PCR analysis of miR-421 in HCMs transfected with miR-NC, miR-421, anti-miR-421 NC or anti-miR-421 (n = 3). g Quantitative RT-PCR analysis of Pink1 in I/R-treated mice and sham mice (n = 10). GAPDH was used as a normalization control. h Pearson correlation analysis of miR-421 and Pink1. All data were from at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
CircPAN3 regulated myocardial I/R injury by targeting miR-421/Pink1 axis-mediated autophagy
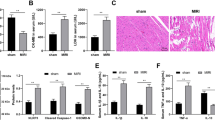
To verify whether Pink1 was the functional target of circPAN3/miR-421 in myocardial I/R injury, mice were infected with adenovirus harbouring circPAN3 or Pink1 siRNA. We found that circPAN3 could obviously alleviate the structural damage of myocardial tissue and myocardial apoptosis caused by I/R treatment; however, the synchronous knockdown of Pink1 completely reversed these effects (Fig. 7a). The reduced myocardial infarct size and autophagy upon circPAN3 overexpression were totally abolished by knockdown of Pink1, suggesting that circPAN3 might regulate autophagy by modulating Pink1 expression (Fig. 7b, c). Pink1 was upregulated upon circPAN3 overexpression and knocked down by Pink1 siRNA (Fig. 7d). Moreover, the effect of circPAN3 overexpression on the protein expression of LC3, Beclin1, P62, Bcl-2, Bax and Caspase 3 was completely abolished by knockdown of Pink1 in mice with I/R injury (Fig. 7e). Taken together, our results demonstrated that circPAN3 might ameliorate myocardial I/R injury by regulating the miR-421/Pink1 axis and Pink1-mediated autophagy.
a H&E and TUNEL staining of heart sections from sham mice and mice treated with I/R, I/R + circPAN3 or I/R + circPAN3 plus Pink1 siRNA (I/R + circPAN3 + siPink1) (n = 10). b TTC staining of heart sections from the indicated mice (n = 10). c The examination of autophagic vacuoles by TEM in heart tissues from the indicated mice (n = 10). d Western blot analysis of Pink1 in the indicated mice. GAPDH was used as a normalization control. e Western blot analysis of LC3II/I, Beclin1, P62, Bax, Bcl-2 and caspase 3 in the indicated mice. GAPDH was used as a normalization control. All data were from at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
Discussion
CVDs are regarded as highly lethal heart diseases worldwide and are the main cause of mortality in many countries [31, 32]. As the most common form of CVD, ischaemic heart diseases, which are characterized by limited blood and oxygen supply to the heart, can lead to extremely fatal conditions and heart attack. Reperfusion is the only effective therapeutic strategy in treating acute ischaemic heart diseases such as severe myocardial infarction; however, it might cause additional I/R injury [33]. Growing evidence has demonstrated that autophagy plays an important role in myocardial I/R injury. For example, autophagy is stimulated in different ways and might play distinct roles in ischaemia and reperfusion [34]. Huang et al. [35] reported that antithrombin III ameliorated I/R injury by suppressing excessive autophagy. A study from Gao et al. [36] showed that TXNIP (thioredoxin-interacting protein)-Redd1 (regulated in development and DNA damage responses 1) contributed to I/R injury by enhancing autophagy. In this study, we are the first to raise the possibility that circPAN3 might ameliorate myocardial I/R injury by inhibiting Pink1-mediated autophagy. Combined with other studies, our results indicate that targeting autophagy in I/R injury might be a potential therapeutic strategy. However, in future investigations are needed to elucidate the direct relationship between autophagy and I/R injury regulated by circPAN3, which was not performed in this study.
As an important modulator in autophagy, circRNAs have attracted much attention from researchers since 2012 owing to their contributions to the regulation of gene expression and disease development [37, 38]. Abnormal expression of circRNAs has been demonstrated to be closely related to the progression and prognosis of various diseases, including carcinomas [39, 40], diabetes [41], kidney diseases [42] and CVDs [43]. CircRNAs generally act as ceRNAs to directly target and absorbs miRNAs to regulate gene expression and play significant roles in diseases such as myocardial I/R injury [28, 44]. CircPAN3, the subject of our investigation, is involved in regulating drug resistance in acute myeloid leukaemia [20] and the self-renewal of intestinal stem cells [19]; however, to the best of our knowledge, its role in myocardial I/R injury has never been reported. In our study, we found for the first time that circPAN3 was downregulated in I/R injury and that its overexpression could ameliorate I/R injury and suppress autophagy. Similar to many other circRNAs targeting miRNAs, circPAN3 could directly target miR-421, a novel target of circPAN3 reported in this study, and act as a miR-421 sponge to regulate I/R injury and autophagy.
miR-421 has been previously reported to act as a crucial modulator in myocardial I/R injury by targeting Pink1 [24, 45]. We found that miR-421 was upregulated upon I/R injury. Overexpression of miR-421 enhanced autophagy and HCM apoptosis in response to H/R treatment, and these effects were reversed by circPAN3 overexpression. Together, previous reports and our data suggest that miR-421 is the functional target of circPAN3 in relieving myocardial I/R injury. As a well-known target of miR-421 and one of the most important regulators in autophagy, Pink1 was confirmed as a direct target of miR-421 and was regulated by circPAN3/miR-421, and knockdown of Pink1 reversed the ameliorating effect of circPAN3 overexpression on myocardial I/R injury in this study. These results demonstrated that circPAN3/miR-421 might regulate myocardial I/R injury by modulating Pink1 and autophagy. However, the detailed regulatory mechanisms of miR-421/Pink1 in I/R injury and autophagy are still unclear and need further investigation.
Based on our results, the circPAN3/miR-421/Pink1 axis could be targeted to treat myocardial I/R injury; however, targeting circPAN3 or miR-421 might result in multiple effects, as these molecules can exert different functions in different systems. Therefore, we still need and plan to investigate the regulatory mechanisms of circPAN3/miR-421 in myocardial I/R injury to provide more reasonable targets for designing therapeutic strategies. Targeting autophagy and Pink1 might be a better option for managing I/R injury.
In summary, in this study, we demonstrated for the first time that circPAN3 might ameliorate myocardial I/R injury by regulating the miR-421/Pink1 axis and inhibiting Pink1-mediated autophagy. Our investigation not only emphasized the important roles of the circPAN3/miR-421/Pink1 axis but also helped elucidate its regulatory mechanism in myocardial I/R injury. More importantly, our study might provide potential targets for developing novel strategies for the treatment of I/R injury.
Change history
22 January 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41374-021-00528-2
References
Mortality GBD, Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459–544.
Kalogeris T, Baines CP, Krenz M, Korthuis RJ. Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol. 2012;298:229–317.
Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J Clin Invest. 2013;123:92–100.
Shimokawa H, Yasuda S. Myocardial ischemia: current concepts and future perspectives. J Cardiol. 2008;52:67–78.
Cohn PF. Treatment of chronic myocardial ischemia: rationale and treatment options. Cardiovasc Drugs Ther. 1998;12(Suppl 3):217–23.
Vishwakarma VK, Upadhyay PK, Gupta GK, Yadav HN. Pathophysiologic role of ischemia reperfusion injury: a review. J Indian Coll Cardiol. 2017;7:97–104.
Zhang W, Xing B, Yang L, Shi J, Zhou X. Icaritin attenuates myocardial ischemia and reperfusion injury via anti-inflammatory and anti-oxidative stress effects in rats. Am J Chin Med. 2015;43:1083–97.
Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–64.
Thorburn A. Apoptosis and autophagy: regulatory connections between two supposedly different processes. Apoptosis. 2008;13:1–9.
Wu D, Jiang H, Chen S, Zhang H. Inhibition of microRNA-101 attenuates hypoxia/reoxygenationinduced apoptosis through induction of autophagy in H9c2 cardiomyocytes. Mol Med Rep. 2015;11:3988–94.
Jiang P, Mizushima N. Autophagy and human diseases. Cell Res. 2014;24:69–79.
Zhang X, Ji J, Yang Y, Zhang J, Shen L. Stathmin1 increases radioresistance by enhancing autophagy in non-small-cell lung cancer cells. Onco Targets Ther. 2016;9:2565–74.
Ouyang F, Huang H, Zhang M, Chen M, Huang H, Huang F, et al. HMGB1 induces apoptosis and EMT in association with increased autophagy following H/R injury in cardiomyocytes. Int J Mol Med. 2016;37:679–89.
Valentim L, Laurence KM, Townsend PA, Carroll CJ, Soond S, Scarabelli TM, et al. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J Mol Cell Cardiol. 2006;40:846–52.
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38.
Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–64.
Zhu P, Zhu X, Wu J, He L, Lu T, Wang Y, et al. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat Immunol. 2019;20:183–94.
Shang J, Chen WM, Liu S, Wang ZH, Wei TN, Chen ZZ, et al. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk Res. 2019;85:106198.
Bittel DC, Kibiryeva N, Marshall JA, O’Brien JE. MicroRNA-421 dysregulation is associated with tetralogy of fallot. Cells. 2014;3:713–23.
Jiang Z, Guo J, Xiao B, Miao Y, Huang R, Li D, et al. Increased expression of miR-421 in human gastric carcinoma and its clinical association. J Gastroenterol. 2010;45:17–23.
Zhou S, Wang B, Hu J, Zhou Y, Jiang M, Wu M, et al. miR-421 is a diagnostic and prognostic marker in patients with osteosarcoma. Tumour Biol. 2016;37:9001–7.
Wang K, Zhou LY, Wang JX, Wang Y, Sun T, Zhao B, et al. E2F1-dependent miR-421 regulates mitochondrial fragmentation and myocardial infarction by targeting Pink1. Nat Commun. 2015;6:7619.
Kang C, Badr MA, Kyrychenko V, Eskelinen EL, Shirokova N. Deficit in PINK1/PARKIN-mediated mitochondrial autophagy at late stages of dystrophic cardiomyopathy. Cardiovasc Res. 2018;114:90–102.
Xu Z, Alloush J, Beck E, Weisleder N. A murine model of myocardial ischemia-reperfusion injury through ligation of the left anterior descending artery. J Vis Exp 2014: 51239. https://doi.org/10.3791/51329.
Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li PF. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci USA. 2009;106:12103–8.
Li M, Ding W, Tariq MA, Chang W, Zhang X, Xu W, et al. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics. 2018;8:5855–69.
Bundgaard A, James AM, Gruszczyk AV, Martin J, Murphy MP, Fago A. Metabolic adaptations during extreme anoxia in the turtle heart and their implications for ischemia-reperfusion injury. Sci Rep. 2019;9:2850.
Yonekawa T, Thorburn A. Autophagy and cell death. Essays Biochem. 2013;55:105–17.
Mc Namara K, Alzubaidi H, Jackson JK. Cardiovascular disease as a leading cause of death: how are pharmacists getting involved? Integr Pharm Res Pract. 2019;8:1–11.
Jones DS, Greene JA. The decline and rise of coronary heart disease: understanding public health catastrophism. Am J Public Health. 2013;103:1207–18.
Yang CF. Clinical manifestations and basic mechanisms of myocardial ischemia/reperfusion injury. Ci Ji Yi Xue Za Zhi. 2018;30:209–15.
Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–22.
Huang KY, Wang JN, Zhou YY, Wu SZ, Tao LY, Peng YP, et al. Antithrombin III alleviates myocardial ischemia/reperfusion injury by inhibiting excessive autophagy in a phosphoinositide 3-kinase/Akt-dependent manner. Front Pharmacol. 2019;10:516.
Gao C, Wang R, Li B, Guo Y, Yin T, Xia Y, et al. TXNIP/Redd1 signaling and excessive autophagy: a novel mechanism of myocardial ischemia/reperfusion injury in mice. Cardiovasc Res. 2019;116:645–57.
Huang R, Zhang Y, Han B, Bai Y, Zhou R, Gan G, et al. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy. 2017;13:1722–41.
Haddad G, Lorenzen JM. Biogenesis and function of circular RNAs in health and in disease. Front Pharmacol. 2019;10:428.
Xie B, Zhao Z, Liu Q, Wang X, Ma Z, Li H. CircRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene. 2019;683:253–61.
Li L, Guo L, Yin G, Yu G, Zhao Y, Pan Y. Upregulation of circular RNA circ_0001721 predicts unfavorable prognosis in osteosarcoma and facilitates cell progression via sponging miR-569 and miR-599. Biomed Pharmacother. 2019;109:226–32.
Fang Y, Wang X, Li W, Han J, Jin J, Su F, et al. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int J Mol Med. 2018;42:1865–74.
Kolling M, Seeger H, Haddad G, Kistler A, Nowak A, Faulhaber-Walter R, et al. The circular RNA ciRs-126 predicts survival in critically ill patients with acute kidney injury. Kidney Int Rep. 2018;3:1144–52.
Zhou LY, Zhai M, Huang Y, Xu S, An T, Wang YH, et al. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death Differ. 2019;26:1299–315.
Wang S, Chen J, Yu W, Deng F. Circular RNA DLGAP4 ameliorates cardiomyocyte apoptosis through regulating BCL2 via targeting miR-143 in myocardial ischemia-reperfusion injury. Int J Cardiol. 2019;279:147.
Chen J, Yu W, Ruan Z, Wang S. TUG1/miR-421/PINK1: a potential mechanism for treating myocardial ischemia-reperfusion injury. Int J Cardiol. 2019;292:197.
Funding
This work was supported by Hunan Provincial Natural Science Foundation Youth Fund (No. 2020JJ5945).
Author information
Authors and Affiliations
Contributions
Guarantor of integrity of the entire study, study concepts, study design and manuscript review: FL. Definition of intellectual content and literature research: S-SB. Experimental studies and manuscript preparation: C-LZ and T-YL. Data acquisition: C-LZ. Data analysis: C-LZ and SAS. Statistical analysis: T-YL. Manuscript editing: SAS.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
All animal procedures were approved by the Institutional Animal Care and Use Committee of Xiangya Hospital of Central South University and conducted in accordance with the National Institutes of Health guidelines.
Consent for publication
The informed consent obtained from study participants.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: There was an error in figure 6
Rights and permissions
About this article
Cite this article
Zhang, CL., Long, TY., Bi, SS. et al. CircPAN3 ameliorates myocardial ischaemia/reperfusion injury by targeting miR-421/Pink1 axis-mediated autophagy suppression. Lab Invest 101, 89–103 (2021). https://doi.org/10.1038/s41374-020-00483-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-020-00483-4
This article is cited by
-
Circular RNA-circPan3 attenuates cardiac hypertrophy via miR-320-3p/HSP20 axis
Cellular & Molecular Biology Letters (2024)
-
The Function of Circular RNAs in Myocardial Ischemia–Reperfusion Injury: Underlying Mechanisms and Therapeutic Advancement
Cardiovascular Drugs and Therapy (2024)
-
The Function and Therapeutic Potential of Circular RNA in Cardiovascular Diseases
Cardiovascular Drugs and Therapy (2023)
-
The circRNA CNEACR regulates necroptosis of cardiomyocytes through Foxa2 suppression
Cell Death & Differentiation (2022)
-
Non-coding RNAs modulate autophagy in myocardial ischemia-reperfusion injury: a systematic review
Journal of Cardiothoracic Surgery (2021)