Abstract
Tyrosine kinase inhibitors (TKIs) are widely accepted as treatment for metastatic clear cell renal cell carcinoma (ccRCC). However, most patients eventually experience disease progression despite TKI treatment, even if the initial response is favorable. To define the underlying mechanism of TKI resistance, 10 TKI-treated metastatic ccRCC cases in which tumor samples were harvested before treatment and immediately after disease progression were examined. Gene expression profiles and copy number variations of matched pre- and post-treatment tumor samples were investigated. Altered biologic characteristics were confirmed in sunitinib-resistant ccRCC cell lines, which were generated by long-term treatment with sunitinib-containing media. Gene transcript levels related to the cell cycle and epithelial-mesenchymal transition (EMT) were significantly upregulated in the treated tumor samples compared with the pre-treatment samples. The mitotic count and sarcomatoid component were significantly increased in treated tumor samples. Alteration of EMT-related genes was also demonstrated in a sunitinib-resistant ccRCC cell line that showed enhanced migration and invasion compared to the parent cell line. siRNA-induced inhibition of EMT-related gene expression significantly suppressed the migration and invasion capacity of TKI-resistant cell lines. The present study shows that both ccRCC cases that progressed after TKI treatment and sunitinib-resistant ccRCC cell lines demonstrated alteration of EMT-related gene expression and enhancement of EMT-related behavior. These results suggest that EMT may explain the aggressive behavior of TKI-resistant ccRCC.
Similar content being viewed by others
Introduction
Clear cell renal cell carcinoma (ccRCC) is the most common histologic subtype of renal cell carcinoma, accounting for ~70% of cases, and is the most lethal malignant neoplasm in the urinary system [1]. Approximately 30–40% of renal cell carcinoma cases are present as metastatic disease, either initially or after curative treatment. Metastatic ccRCC is considered to have poor prognosis, with a 5-year survival rate of only 8% [2]. Metastatic ccRCC patients are treated with tyrosine kinase inhibitors (TKIs), including sunitinib, pazopanib, sorafenib, and axitinib, as first-line agents. A clinically beneficial response to TKI treatment has been observed in several clinical trials, with an objective response rate of up to 31% [2,3,4,5]; however, nearly all patients develop resistance to TKI therapy, the mechanism of which remains largely unknown [6].
Epithelial-to-mesenchymal transition (EMT) is driven by upregulation of specific transcription factors that activate mesenchymal genes and repress epithelial genes, including Twist1/2, Snail, Slug, and ZEB1/2 [7, 8]. EMT is integral to embryonal development, tissue regeneration, and fibrosis [9]. It also plays an important role in cancer progression and is involved in migration and invasion of tumor cells, suppression of senescence and apoptosis, and resistance to radiotherapy and chemotherapy [10,11,12]. Induction of EMT generates CD44-expressing cancer stem cells in various cancers [13,14,15]. In renal cell carcinoma, sarcomatoid transformation, which is a histologic manifestation of EMT, has been associated with poor response to TKI therapy [16,17,18]. However, the role of EMT in the development of TKI resistance in ccRCC remains to be elucidated [19, 20].
Materials and methods
Patients
This study was approved by the Asan Medical Center Institutional Review Board (2012-0788) and the procedures involving human subjects were carried out in accordance with the revised Helsinki Declaration in 1983. Initially, 553 cases of recurrent or metastatic RCC patients treated with vascular endothelial growth factor receptor (VEGFR)-TKI at the Asan Medical Center, Seoul, Republic of Korea, from 1997 to 2013 were retrieved. To define the mechanism of resistance to TKI in ccRCC, we searched for cases with clinical signs of progression despite TKI treatment, then further extracted those cases with matched pairs of pre- and post-treatment tumor samples available, and with the post-treatment tumor samples that were obtained within a month after the progression had been recognized clinically. Finally, 10 patients that fulfilled the selection criteria were included in the analysis.
Response to VEGFR-TKI was assessed according to the Revised Response Evaluation Criteria in Solid Tumors guidelines (version 1.1) [21]. Patients showing complete response, partial response, or ≥24 weeks stable disease in response to TKI treatment were defined as the clinical benefit group, whereas those showing <24 weeks stable disease or progressive disease were defined as the clinical non-benefit group by an oncologist (J.L.) at this institution [22].
Patient medical records were reviewed for characteristics including age, sex, tumor stage during TKI treatment, and outcome. Pathologic materials were reviewed by three uropathologists (H.S.H., H.G., and Y.M.C.) according to the 2016 World Health Organization (WHO) Tumor Classification, graded according to the ISUP grading system, and staged according to the American Joint Committee on Cancer (AJCC) Staging System, 8th edition [23, 24].
Total RNA expression profiling
The RNAs of the 20 matched pairs of pre- and post-treatment formalin-fixed paraffin-embedded (FFPE) tumor samples derived from 10 enrolled patients were extracted and subjected to expression profiling. Briefly, the tumor areas of the samples were identified on the FFPE tissue block by two uropathologists (H.G. and Y.M.C.) and cut from the tissue section. After deparaffinization, RNA from tumor tissues was extracted using the Ambion RecoverAll Total Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol. Then, 50 ng of extracted RNA per tumor sample was analyzed for total RNA expression profiling using the Affymetrix Human Transcriptome Array 2.0 platform (Affymetrix, Santa Clara, CA). The data were normalized using Transcriptome Analysis Center version 3.0 (Affymetrix) by default option. The probes were annotated using the chip annotation file for Human Transcriptome Array 2.0 platform provided on the Broad Institute website (http://software.broadinstitute.org/gsea/index.jsp) (Broad Institute, Cambridge, MA).
Copy number variation analysis
Copy number variations (CNV) were also evaluated in the 10 pairs of pre- and post-treatment tumor samples. DNA was extracted from FFPE tumor samples using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). Then, 50 ng of extracted DNA per individual sample was analyzed using the Affymetrix OncoScanⓇ FFPE Assay Kit. The data was analyzed using Nexus Express Software for OncoScan version 3.1 (BioDiscovery, El Segundo, CA).
Differentially expressed genes and gene set analysis
Differential gene expression between paired pre- and post-treatment tumor samples was evaluated by the empirical Bayes moderated paired t-test using R package limma [25]. We set the positive criterion for differential expression as an unadjusted P-value < 0.05 because of the small number of cases.
For gene set analysis, both over-representation and gene set enrichment analysis (GSEA) were performed using R package clusterProfiler and GSEA Java program provided by the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp), respectively [26, 27]. For GSEA, the GSEAPreranked tool was used because it supports pairwise comparison while the conventional GSEA method does not. The t-statistics for the genes calculated during DEG analysis were used as input data during GSEAPreranked analysis. False discovery rate (FDR) < 0.05 was considered as statistically significant for the gene set analysis.
Tissue microarray construction and immunohistochemistry
A tissue microarray was constructed using two representative cores 1.5 mm in diameter from FFPE tumor tissues and was employed in immunohistochemical staining.
Immunohistochemistry was carried out using an automated staining system (BenchMark XT, Ventana Medical Systems, Tucson, AZ) according to the manufacturer’s instructions. The primary antibodies used are listed in Supplementary Table 1.
Generation of sunitinib-resistant renal cell carcinoma cell lines
Two renal cell carcinoma cell lines, ACHN and Caki-1, were grown in RPMI 1640 supplemented with 10% fetal bovine serum in a humidified atmosphere containing 5% CO2 at 37 °C. To generate TKI-resistant renal cell carcinoma cell lines, ACHN and Caki-1 were treated with sunitinib dissolved in culture media at 2.5 and 2.0 μM, respectively, for 6 months. Sunitinib resistance was confirmed by cell viability assay as described below.
Cell viability assay
The cells were seeded onto 96-well plates and incubated with 1 mg/mL 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyl-tetrazolium bromide (MTT) for 4 h. After the removal of MTT, dimethyl sulfoxide (DMSO) was added to each well and the absorbance was measured at 540 nm. Each experiment was performed in triplicate and repeated three times for reliable comparison.
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the cultured ACHN and Caki-1 cell lines. Complementary DNA (cDNA) was synthesized from 300 ng RNA using PrimeScript RT Master Mix (Takara Bio Inc., Shiga, Japan). qRT-PCR was performed using the Applied Biosystems 7500 Fast Real-time PCR System (Thermo Fisher Scientific) according to the manufacturer’s protocol. The primer sequences were designed using Primer3 software (http://primer3.ut.ee) (Supplementary Table 2). Gene expression is reported as the ΔCt value [28]. The ACTB gene was used as an endogenous control. The procedures were performed in triplicate and repeated three times.
RNA interference
siRNAs targeting CLDN1 (SI03206336, Qiagen), CD44 (L-009999-00-0005, Dharmacon, Lafayette, CO), SNAI2 (L-017386-00-0005, Dharmacon), and TWIST2 (L-012862-02-0005, Dharmacon) genes were purchased from the indicated manufacturers. The negative control (NCT) siRNAs were Stealth RNAiTM siRNAs purchased from Thermo Fisher Scientific. Lipofectamine® 2000 (Thermo Fisher Scientific) was used for siRNA transfection. Gene expression, cell viability, and wound healing and invasion assays were evaluated at 48 h post-transfection.
Western blotting
Total protein was extracted from the cultured cells using RIPA lysis and extraction buffer (Thermo Fisher Scientific). The proteins extracts were subjected to SDS-PAGE on a 12% polyacrylamide gel for 2 h. After electrophoresis, protein was transferred onto an iBlot 2 polyvinylidene difluoride membrane with the iBlot 2 dry blotting system (Thermo Fisher Scientific). Then, the membrane was blocked with 5% skim milk for 1 h and incubated with primary antibody overnight at 4 °C, followed by incubation with secondary antibody conjugated to horseradish peroxidase. The immunoreactivity signal was developed using SuperSignal West Dura Substrate (Thermo Fisher Scientific). β-actin was used as a control for protein loading. The dilution levels of primary antibodies are listed in Supplementary Table 1.
Wound healing assay and Matrigel invasion assay
The wound healing assay and Matrigel invasion assay were performed to evaluate cell migration capacity and invasiveness, respectively. For the wound healing assay, cells were seeded to reach ~80% confluence as a monolayer after 24 h of growth. The cell monolayer was scratched using a 10 μL pipette tip across the center. After washing, cells were incubated for 48 h. During the incubation, microphotographs of the cell monolayer were taken at 0, 16, 24, and 48 h post-scratch. The unoccupied areas were measured at the indicated times using the ImageJ version 1.50i with MRI wound healing tool macro (http://dev.mri.cnrs.fr/projects/imagej-macros/wiki/Wound_Healing_Tool) (National Institute of Health, Bethesda, MD).
For Matrigel invasion assay, Matrigel basement membrane matrix (Corning Inc., Corning, NY) was coated onto the transwells of Falcon® Permeable Supports (Corning). A total of 2.5 × 105 cells were seeded into each transwell. After 24 h, photomicrographs of the lower wells were taken to determine the number penetrated cells.
Statistical analysis
Pairwise comparisons of continuous variables were carried out by the Wilcoxon signed-rank test. For pairwise comparison of ordinal categorical variables, cumulative link mixed models regarding the case number as a random variable were fitted using the clmm function in R package ordinal. The statistical significance of the variable was calculated using the Anova.clm function in R package RVAideMemoire. Two-sided P-value < 0.05 was considered statistically significant.
Results
Clinical characteristics of patients with TKI resistance
The clinical and pathologic characteristics of the 10 patients examined in this study are listed in Table 1. The median age at the start of TKI treatment was 53.5 years (range, 40–66 years). Most of the patients were male (male:female ratio = 4:1). Six patients presented with stage IV disease at the time of the diagnosis; the other four patients were treated with TKI due to post-nephrectomy recurrence. Seven patients (70%) were treated with sunitinib; the other three received pazopanib. After TKI treatment, eight (80%) achieved remission status and were regarded as the clinical benefit group, with complete and partial remission in two and six patients, respectively. The remaining two patients displayed stable disease lasting < 24 weeks and were deemed the clinical non-benefit group. Despite treatment, all submitted patients exhibited disease progression at a median time of 13.5 months (range, 1–70 months) after TKI treatment. Eventually all patients died of the disease in a median time of 24.5 months (range, 5–96 months) after TKI treatment.
Microarray analysis reveals differentially expressed genes and enriched pathways in TKI-resistant post-treatment samples
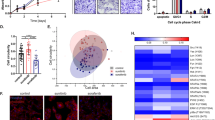
Microarray analysis of pre- and post-TKI ccRCC samples revealed 2944 differentially expressed transcripts. The number of annotated transcripts was 975, of which 715 transcripts were upregulated and 260 were downregulated (Fig. 1a). Excluding the overlapping gene names of the differentially expressed transcripts, 675 upregulated and 250 downregulated genes were identified. The top 30 upregulated and downregulated transcripts are presented in Supplementary Tables 3 and 4, respectively.
Microarray analysis between pre- and post-treatment ccRCC tumor samples. a Gene expression heatmap showing differentially expressed transcripts between pre- (green) and post-treatment (red) samples. Upregulated transcripts in post-treatment tumor samples include cell cycle regulators (CDK2 and CDK4) and EMT-associated genes (CD44, SDC4, CLDN1, SNAI2, and TWIST2). Macrophage markers (CD68 and CD163) and some cytokine (CXCL1) genes were also upregulated. b Enrichment plots of gene set enrichment analysis (GSEA) revealing significant enrichments for cell cycle regulators and EMT gene sets
Over-representation analyses for hallmark (H) and canonical pathway (C2cp) gene sets were performed using the gene names of the upregulated and downregulated transcripts (Table 2). The significantly enriched gene sets for the upregulated genes in post-treatment samples included mammalian targets of the rapamycin (mTOR) signaling pathway, cell proliferation, oxidative phosphorylation, and EMT-associated genes including CD44, SNAI2, TWIST, and CLDN1 (Fig. 1a). There was no significant difference in the expression of other EMT-related genes, such as ZEB1, ZEB2, SNAI1, TWIST1, and CDH1 (data not shown). GSEA also displayed significant enrichment for similar gene sets (Fig. 1b, Supplementary Table 5).
Gene expression is not altered by copy number alteration in TKI-resistant post-treatment samples
We evaluated the difference in copy number variation between the pre- and post-treatment ccRCC samples (Supplementary Figure 1). Both the pre- and post-treatment samples showed high frequency of copy number loss of chromosome 3p, which is one of the most common genetic alterations of ccRCC [29]. Compared to the pre-treatment samples, post-treatment samples displayed significantly increased copy number gains at the chromosome regions 7q36.3, 8q23.3, and 12q13.13, which encompass the PTPRN2, CSMD3, and ATF7 genes, respectively. However, according to the microarray data, these genes were not upregulated in post-treatment tumor samples. These results suggest that these copy number alterations were not directly associated with gene expression elevation, and may play insignificant roles in TKI resistance [30].
Pairwise comparisons of the pathologic features confirm increased proliferative activity and EMT in post-treatment ccRCC samples
Compared to the pre-treatment samples, post-treatment samples revealed an increased proportion of sarcomatoid spindle cell component and mitotic counts (P = 0.036 and P = 0.009, respectively) (Fig. 2a, b). The ISUP nuclear grades tended to be higher in post-treatment samples, but this relationship was not statistically significant (P = 0.103). The proportion of tumor necrosis was not significantly different (data not shown).
Histopathologic and immunohistochemical features of pre- and post-treatment ccRCC samples. a Representative photomicrographs of pre- and post-treatment tumor samples of Case No. 4. Contrary to the pre-treatment tumor showing typical histologic features of ccRCC, the post-treatment tumor exhibits a sheet-like arrangement of spindle tumor cells with brisk mitotic activity (black arrow). b Pairwise comparison of the proportion of sarcomatoid component and mitotic activities of pre-treatment (PreTx) and post-treatment (PostTx) tumor samples showing significant elevation of both parameters in post-treatment tumor samples (paired t-test, P < 0.05). c Representative photomicrographs of CD44, Slug, Claudin-1, and Twist-2 immunohistochemical staining. Images of both pre-TKI treatment (left) and post-TKI treatment (right) have been shown. d Heatmap of the immunoreactivity scores for CD44, Slug, Claudin-1, and Twist-2 showing a significant upregulation of CD44, Slug, and Twist-2 proteins. The statistical significance of the difference between the immunoreactivity scores of the pre- and post-treatment tumor samples was evaluated using the cumulative link mixed model. The criterion for the scoring is indicated at the bottom
Since we previously determined that sarcomatoid features are a predictive marker for TKI responsiveness in ccRCC [18], we decided to focus on EMT for subsequent experiments. The increased expression of EMT-related genes determined by microarray was confirmed by immunohistochemistry in the pre- and post-treatment samples. The expression levels of CD44, Slug, and Twist-2 were upregulated in the majority of post-treatment samples (P = 0.020, P < 0.001, and P = 0.001, respectively) (Fig. 2c, d, and Supplementary Table 6); however, a proper statistical comparison was limited due to the small number of cases. Claudin-1 expression was elevated in some, but not all post-treatment samples (Fig. 2d). In line with the results of mRNA expression, no significant differences were noted in the protein expression levels of ZEB-1, Snail, and E-cadherin (Supplementary Figure 2).
Generation of sunitinib-resistant renal cell carcinoma cell lines
Sunitinib-resistant renal cell carcinoma cell lines designated as ACHN/suR and Caki-1/suR were generated by long-term exposure to sunitinib at concentrations of 2.5 μM and 2.0 μM, respectively, for 6 months. Compared to the ACHN (ACHN/P) and Caki-1 (Caki-1/P) parental cell lines, ACHN/suR and Caki-1/suR revealed higher viability at various concentrations of sunitinib, confirming their resistance (Fig. 3a).
a Results of cell viability tests for sunitinib-resistant and parent ccRCC cell lines, ACHN and Caki-1. Compared to the parent cell lines (dotted lines), sunitinib-resistant cell lines (solid lines) exhibited superior survival rates in various concentration of sunitinib. b Representative microscopic images of cells from parental (P) cell lines and the sunitinib-resistant (suR) ACHN and Caki-1 cell lines. c Comparison of the mRNA expression levels of EMT-related genes in the sunitinib-resistant and parent ccRCC cell lines obtained by qRT-PCR analysis. ACHN/suR displayed a significantly increased expression of the SNAI1, SNAI2, TWIST1, and TWIST2 genes, compared to the ACHN/P cell line. In the Caki-1/suR cell line, CLDN1, CD44, ZEB1, ZEB2, and TWIST1 expression levels were significantly elevated. CDH1 expressions were constantly downregulated in both the ACHN/suR and Caki-1/suR cell lines. d Results of western blotting of the sunitinib-resistant and parent cell line samples. The protein expression levels of CD44, Claudin-1, Snail, Slug, and Twist-2 were increased in the sunitinib-resistant cell lines. Although the ZEB-1 expression level was not significantly altered, the E-cadherin expression level was consistently decreased in both the ACHN/suR and Caki-1/suR cell lines
Sunitinib-resistant renal cell carcinoma cell lines show increased expression of EMT-related genes
Compared to the cells of the parental cell lines, ACHN/suR and Caki-1/suR cells were spindle shaped and exhibited a projection of elongated foot processes (Fig. 3b). To confirm EMT-related gene upregulation in TKI-resistant ccRCC samples, expression of the EMT-associated genes CD44, SNAI2, CLDN1, and TWIST2, was examined in the parental and resistant cell lines. Expression levels of known EMT-related genes, ZEB1, ZEB2, SNAI1, and TWIST1, were also evaluated. TWIST1 mRNA was overexpressed both in TKI-resistant cell lines. SNAI1, SNAI2, and TWIST2 mRNA expression levels were increased in ACHN/suR, whereas CLDN1, CD44, ZEB1, and ZEB2 mRNAs were overexpressed in Caki-1/suR (Fig. 3c). Nevertheless, protein expression levels of claudin-1, CD44, Snail, Slug, and Twist-2 were increased in both ACHN/suR and Caki-1/suR (Fig. 3d). Although there was a significant alteration of the mRNA expression level, ZEB-1 protein expression level was not different in the western blotting analysis. mRNA and protein expression levels of CDH1/E-cadherin were downregulated in both ACHN/suR and Caki-1/suR, compared to their parent cell lines (Fig. 3c, d). These results suggest that EMT phenomenon occurred consistently in TKI-resistant ccRCC samples, but this may be regulated by different EMT-related genes across the samples.
Sunitinib-resistant renal cell carcinoma cell lines show enhanced EMT-related behaviors
To examine the migration ability of TKI-resistant ccRCC, ACHN/suR, and Caki-1/suR were subjected to the wound healing assay. Compared to the parental cell lines, both ACHN/suR and Caki-1/suR exhibited increased capability to fill the cleft generated by scratching (Fig. 4c). In the Matrigel invasion assay, the number of cells penetrating the Matrigel membrane was also increased in ACHN/suR and Caki-1/suR compared to the corresponding parent cell lines (Fig. 4d).
a siRNA transfection successfully depleted target mRNAs of CD44 and TWIST2 in sunitinib-resistant ACHN (ACHN/suR) and CLDN1 and SNAI2 in Caki-1 (Caki-1/suR) cell lines. b mRNA depletion of CD44, TWIST2, and SNAI2 did not significantly affect cell viability except CLDN1 in Caki-1, which significantly reduced cell viability similar to that of the parent cell line. c Results of scratch test in sunitinib-resistant cell lines with or without siRNA inhibition. Compared to the parent cell lines, both sunitinib-resistant cell lines filled the central void area faster than the parent cell lines (gray line plots). This ability was successfully inhibited by siRNA-induced depletion of EMT-associated genes. d Results of Matrigel invasion assay showing the significantly increased number of permeating cells in the sunitinib-resistant cell lines, compared to the case for their parent cell lines. This invasive capacity was also decreased by inhibition of EMT-related genes by siRNA
Inhibition of EMT-related genes suppress EMT-related behavior
To examine the effect of EMT-related genes in TKI-resistant ccRCC, knockdown of CD44, TWIST2, CLDN1, and SNAI2 genes was achieved using siRNA transfection. The expression of CD44 and TWIST2 was successfully inhibited in ACHN/suR, whereas expression of CLDN1 and SNAI2 was suppressed in Caki-1/suR (Fig. 4a).
When the expression of EMT-related genes was inhibited, cell survival tended to be decreased in ACHN/suR and Caki-1/suR, but the difference was statistically significant only in CLDN1-inhibited Caki-1/suR (Fig. 4b). In contrast, inhibition of EMT-related genes markedly decreased the migration rates of ACHN/suR and Caki-1/suR in the wound healing assay (Fig. 4c). Furthermore, invasion capacity was also suppressed in ACHN/suR and Caki-1/suR by inhibition of EMT-related genes (Fig. 4d).
Discussion
This study demonstrates that, compared to pre-treatment ccRCC, TKI-resistant post-ccRCC tumors had increased sarcomatoid features and differential gene expression with upregulation of EMT-related genes CD44, CLDN1, SNAI2, and TWIST2. Furthermore, the induction of TKI resistance in ccRCC cell lines increased expression of EMT-related genes with enhanced migration and invasion capacity, which was suppressed by inhibition of EMT-related genes. Therefore, these results suggest that the aggressive behavior of TKI resistance is associated with EMT.
Several studies using PDX models have suggested that sphingosine-1-phosphate or mitogen-activated protein kinase pathways may contribute to TKI resistance in renal cell carcinoma [31, 32]. The PDX model has been known to mimic biologic behaviors of human malignancies, but it may not entirely reproduce the human tumor microenvironment because it cannot recapitulate the antineoplastic immune reaction and tumor interaction with human stromal cells [33]. Furthermore, the pathways implicated in prior studies have not been validated in patient samples. In the present study, in silico analysis of matched-pair tumor samples of patients were carried out, revealing the significant enrichments of EMT, cell proliferation-associated signatures, and mTOR pathway in ccRCC patients with TKI resistance, rather than the pathways stated above. It is shown in the present study that of these pathways, EMT has been suggested as one of the mechanisms underlying the development of resistance to chemotherapeutic agents in non-small cell lung cancer and breast cancer [34, 35]. Although EMT has not been intensively studied in renal cell carcinoma, several reports have implicated the role of EMT in TKI resistance in renal cell carcinoma [36]. Therefore, it can be postulated that the activation of EMT may be an important factor in TKI resistance of ccRCC, which needs to be confirmed in further studies.
EMT response in post-TKI treatment ccRCC is not well studied [20, 37], because post-treatment tumor tissue is rarely biopsied in practice and EMT may be reversible. Furthermore, contrary to conventional cancer therapies that mainly suppress cancer cell proliferation, TKI treatment in ccRCC is known to target the pro-angiogenic signaling pathway of tumor-associated endothelial cells rather than the tumor cells themselves [38]. In this context, angiogenic escape and/or switching have been widely accepted as the major mechanisms underlying TKI resistance [19, 39]. However, there is a report showing that sunitinib may also inhibit renal cell carcinoma cell growth in vivo at a clinically actionable dose [40], which suggests a role of TKI cytotoxicity in the control ccRCC tumor cells. There are few reports implicating the role of EMT in TKI resistance in renal cell carcinoma, but these studies used renal cell carcinoma cell lines and/or patient-derived xenograft (PDX) models, and their findings have not been clinically validated in patient samples [20, 36, 41]. In the present study, using post-TKI treatment tumor samples biopsied immediately after the development of TKI resistance and tumor progression, we found that EMT features were increased in ccRCC patients with upregulation of CD44, CLDN1, SNAI2, and TWIST2. Although significant E-cadherin downregulation was not shown in post-treatment tissue samples, which may be related to the essentially low E-cadherin expression in ccRCC [42, 43], an increased sarcomatoid component in the TKI-resistant tumor samples implies an upregulated EMT response. Moreover, this finding was supported by in vitro studies showing the downregulation of E-cadherin, increased invasion capacity, and upregulation of some EMT-related genes in sunitinib-resistant renal cell carcinoma cell lines. Although mRNA levels of CD44 in ACHN/suR and SNAI2 and TWIST2 in Caki-1/suR were not significantly elevated, their protein expression levels were elevated, which could be due to post-translational modification of these proteins [44, 45]. The invasion capacity of TKI-resistant ccRCC cells was decreased by the depletion of these genes. Our findings suggest that EMT is an important mechanism in TKI-resistant ccRCC and may be a novel therapeutic target for TKI resistance.
CD44 is transmembrane glycoprotein that participates in various biologic processes including cell adhesion, angiogenesis, tumor development, and drug resistance [46, 47]. In renal cell carcinoma, CD44 expression has been associated with poor prognosis, cancer cell invasion, and resistance to TKI treatment, which are consistent with our results showing elevated expression of CD44 in TKI-resistant ccRCC both in vitro and in vivo [48, 49]. In our analyses of TKI-resistant ccRCC cell lines, CD44 depletion was found to significantly decrease cell invasiveness, which confirms the role of cell invasion in TKI-resistant ccRCC. Slug and Twist-2, members of the SNAIL and basic helix-loop-helix families of transcription factors, respectively, are known to promote EMT [8]. Several studies have indicated that Slug and Twist-2 contribute to drug resistance in various human malignancies, including renal cell carcinoma [50,51,52]. The present study indicates that these transcription factors were upregulated in TKI-resistant ccRCC samples. siRNA-mediated depletion of Slug and Twist-2 induced reduction of invasion capacity but failed to inhibit cell proliferation. This is likely because the proliferative capacity was dependent upon other signaling pathways, such as cell cycle or mTOR pathways. Hence, dual inhibition of these pathways may exert a synergistic effect to overcome TKI resistance.
Claudin-1 is a component of tight junctions, which are one of the major constituents involved in cell-to-cell adhesion of epithelial cells [53]. The role of claudin-1 in tumor biology is currently under debate; several studies have suggested that it has a tumor suppressive function [54, 55], while others implicate its role in EMT induction [56,57,58]. In renal cell carcinoma, claudin-1 expression has been associated with an increased risk of postoperative distant metastasis [59] and poor prognosis [60], but its role in EMT induction and drug resistance in renal cell carcinoma has not been elucidated. The present study suggests that claudin-1 expression may be associated with TKI resistance in both clinical and experimental settings, which may be overcome by claudin-1 downregulation. These results indicate the potential of claudin-1 as a prognostic marker and therapeutic target for patients with renal cell carcinoma undergoing TKI treatment [61], which should be researched in future studies.
The expressions of the EMT-associated genes analyzed in the study varied across the TKI-resistant patient samples and cell lines. Although the phenomenon should be clarified further, it could be explained by the fact that EMT represents a reversible phenotype and is controlled by various cellular signaling pathways and posttranscriptional modifications across individual tumors in TKI-resistant ccRCC [8].
The present study has several limitations. First, we could not perform validation of gene expression alterations identified by microarray experiments because of the limited size of biopsy specimens, which were the major tissue type of the enrolled cases. Additionally, in DEG analysis, because of the small number of the submitted cases, we used an unadjusted P-value, which is not a conventional analytical method [62]. However, we showed that immunohistochemical intensities of the proteins of these genes were increased in post-treatment tumor samples with statistical significance for three of four genes, and these genes were also upregulated in sunitinib-resistant renal cell carcinoma cell lines. Furthermore, downregulation of EMT-related gene expression suppressed EMT-related biologic behavior, and even significantly affected cell viability for some genes. Therefore, although the DEG analysis method was unconventional, the EMT-related genes listed above may have a crucial role in TKI resistance of ccRCC.
In conclusion, EMT is consistently observed in ccRCC with TKI resistance. Modulation of several EMT-associated genes may alter the metastatic potential and/or TKI susceptibility in TKI-resistant ccRCC patients, which implicates it as a therapeutic strategy.
References
Moch H, Linehan WM, Bonsib SM, Reuter VE, Delahunt B, Srigley JR, et al. Clear cell renal cell carcinoma. In: Moch H, Humphrey PA, Ulbright TM, Reuter VE, editors. WHO classification of tumors of the urinary system and male genital organs. Lyon: IARC; 2015. p. 18–21.
Choueiri TK, Motzer RJ. Systemic therapy for metastatic renal-cell carcinoma. N Engl J Med. 2017;376:354–66.
Escudier B, Eisen T, Stadler WM, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356:125–34.
Gore ME, Szczylik C, Porta C, et al. Final results from the large sunitinib global expanded-access trial in metastatic renal cell carcinoma. Br J Cancer. 2015;113:12–19.
Motzer RJ, Hutson TE, Tomczak P, et al. Overall survival and updated results for sunitinib compared with interferon alfa in patients with metastatic renal cell carcinoma. J Clin Oncol. 2009;27:3584–90.
Griffioen AW, Mans LA, de Graaf AMA, et al. Rapid angiogenesis onset after discontinuation of sunitinib treatment of renal cell carcinoma patients. Clin Cancer Res. 2012;18:3961–71.
Sui H, Zhu L, Deng W, et al. Epithelial-mesenchymal transition and drug resistance: role, molecular mechanisms, and therapeutic strategies. Oncol Res Treat. 2014;37:584–9.
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96.
Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8.
Du B, Shim JS. Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules. 2016;21:E965.
Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–51.
De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110.
Bhattacharya R, Mitra T, Ray Chaudhuri S, et al. Mesenchymal splice isoform of CD44 (CD44s) promotes EMT/invasion and imparts stem-like properties to ovarian cancer cells. J Cell Biochem. 2018;119:3373–83.
Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15.
Miyazaki H, Takahashi RU, Prieto-Vila M, et al. CD44 exerts a functional role during EMT induction in cisplatin-resistant head and neck cancer cells. Oncotarget. 2018;9:10029–41.
He H, Magi-Galluzzi C. Epithelial-to-mesenchymal transition in renal neoplasms. Adv Anat Pathol. 2014;21:174–80.
Molina AM, Tickoo SK, Ishill N, et al. Sarcomatoid-variant renal cell carcinoma: treatment outcome and survival in advanced disease. Am J Clin Oncol. 2011;34:454–9.
Park JY, Lee JL, Baek S, et al. Sarcomatoid features, necrosis, and grade are prognostic factors in metastatic clear cell renal cell carcinoma with vascular endothelial growth factor-targeted therapy. Hum Pathol. 2014;45:1437–44.
Bielecka ZF, Czarnecka AM, Solarek W, et al. Mechanisms of acquired resistance to tyrosine kinase inhibitors in clear-cell renal cell carcinoma (ccRCC). Curr Signal Transduct Ther. 2014;8:218–28.
Hammers HJ, Verheul HM, Salumbides B, et al. Reversible epithelial to mesenchymal transition and acquired resistance to sunitinib in patients with renal cell carcinoma: evidence from a xenograft study. Mol Cancer Ther. 2010;9:1525–35.
Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumors: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47.
Di Leo A, Jerusalem G, Petruzelka L, et al. Results of the CONFIRM phase III trial comparing fulvestrant 250 mg with fulvestrant 500 mg in postmenopausal women with estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2010;28:4594–4600.
Humphrey PA, Ulbright TM, Reuter VE, et al. WHO classification of tumors of the urinary system and male genital organs. 4th ed. Lyon: International Agency for Research on Cancer; 2016.
Edge SB. American Joint Committee on Cancer. AJCC cancer staging manual. 8th ed. New York: Springer; 2017.
Smyth GK. limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, et al., editors. Bioinformatics and computational biology solutions using R and bioconductor statistics for biology and health. New York: Springer; 2005. p. 397–420.
Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45.
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49.
Myhre S, Lingjaerde OC, Hennessy BT, et al. Influence of DNA copy number and mRNA levels on the expression of breast cancer related proteins. Mol Oncol. 2013;7:704–18.
Diaz-Montero CM, Mao FJ, Barnard J, et al. MEK inhibition abrogates sunitinib resistance in a renal cell carcinoma patient-derived xenograft model. Br J Cancer. 2016;115:920–8.
Zhang L, Wang X, Bullock AJ, et al. Anti-S1P antibody as a novel therapeutic strategy for VEGFR TKI-resistant renal cancer. Clin Cancer Res. 2015;21:1925–34.
Jung J, Seol HS, Chang S. The generation and application of patient-derived xenograft model for cancer research. Cancer Res Treat. 2018;50:1–10.
Huang J, Li H, Ren G. Epithelial-mesenchymal transition and drug resistance in breast cancer (Review). Int J Oncol. 2015;47:840–8.
Nurwidya F, Takahashi F, Murakami A, et al. Epithelial mesenchymal transition in drug resistance and metastasis of lung cancer. Cancer Res Treat. 2012;44:151–6.
Zhou L, Liu XD, Sun M, et al. Targeting MET and AXL overcomes resistance to sunitinib therapy in renal cell carcinoma. Oncogene. 2016;35:2687–97.
Savagner P. The epithelial-mesenchymal transition (EMT) phenomenon. Ann Oncol. 2010;21(Suppl 7):vii89–92.
Huang D, Ding Y, Li Y, et al. Sunitinib acts primarily on tumor endothelium rather than tumor cells to inhibit the growth of renal cell carcinoma. Cancer Res. 2010;70:1053–62.
Porta C, Paglino C, Imarisio I, et al. Changes in circulating pro-angiogenic cytokines, other than VEGF, before progression to sunitinib therapy in advanced renal cell carcinoma patients. Oncology. 2013;84:115–22.
Gotink KJ, Broxterman HJ, Labots M, et al. Lysosomal sequestration of sunitinib: a novel mechanism of drug resistance. Clin Cancer Res. 2011;17:7337–46.
Mizumoto A, Yamamoto K, Nakayama Y, et al. Induction of epithelial-mesenchymal transition via activation of epidermal growth factor receptor contributes to sunitinib resistance in human renal cell carcinoma cell lines. J Pharmacol Exp Ther. 2015;355:152–8.
Langner C, Ratschek M, Rehak P, et al. Expression of MUC1 (EMA) and E-cadherin in renal cell carcinoma: a systematic immunohistochemical analysis of 188 cases. Mod Pathol. 2004;17:180–8.
Mikami S, Katsube K, Oya M, et al. Expression of Snail and Slug in renal cell carcinoma: E-cadherin repressor Snail is associated with cancer invasion and prognosis. Lab Invest. 2011;91:1443–58.
Diaz VM, Vinas-Castells R, Garcia, de Herreros A. Regulation of the protein stability of EMT transcription factors. Cell Adh Migr. 2014;8:418–28.
Jordan AR, Racine RR, Hennig MJ, et al. the role of CD44 in disease pathophysiology and targeted treatment. Front Immunol. 2015;6:182.
Xu H, Tian Y, Yuan X, et al. The role of CD44 in epithelial-mesenchymal transition and cancer development. Onco Targets Ther. 2015;8:3783–92.
Cain JW, Hauptschein RS, Stewart JK, et al. Identification of CD44 as a surface biomarker for drug resistance by surface proteome signature technology. Mol Cancer Res. 2011;9:637–47.
Lim SD, Young AN, Paner GP, et al. Prognostic role of CD44 cell adhesion molecule expression in primary and metastatic renal cell carcinoma: a clinicopathologic study of 125 cases. Virchows Arch. 2008;452:49–55.
Mikami S, Mizuno R, Kosaka T, et al. Expression of TNF-alpha and CD44 is implicated in poor prognosis, cancer cell invasion, metastasis and resistance to the sunitinib treatment in clear cell renal cell carcinomas. Int J Cancer. 2015;136:1504–14.
Haslehurst AM, Koti M, Dharsee M, et al. EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer. 2012;12:91.
Wang T, Li Y, Tuerhanjiang A, et al. Twist2 contributes to cisplatin-resistance of ovarian cancer through the AKT/GSK-3beta signaling pathway. Oncol Lett. 2014;7:1102–8.
Zhang HJ, Tao J, Sheng L, et al. Twist2 promotes kidney cancer cell proliferation and invasion by regulating ITGA6 and CD44 expression in the ECM-receptor interaction pathway. Onco Targets Ther. 2016;9:1801–12.
Morita K, Furuse M, Fujimoto K, et al. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA. 1999;96:511–6.
Chao YC, Pan SH, Yang SC, et al. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am J Respir Crit Care Med. 2009;179:123–33.
Higashi Y, Suzuki S, Sakaguchi T, et al. Loss of claudin-1 expression correlates with malignancy of hepatocellular carcinoma. J Surg Res. 2007;139:68–76.
Dhawan P, Singh AB, Deane NG, et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005;115:1765–76.
Huang J, Zhang L, He C, et al. Claudin-1 enhances tumor proliferation and metastasis by regulating cell anoikis in gastric cancer. Oncotarget. 2015;6:1652–65.
Suh Y, Yoon CH, Kim RK, et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene. 2013;32:4873–82.
Shin HI, Kim BH, Chang HS, et al. Expression of claudin-1 and -7 in clear cell renal cell carcinoma and its clinical significance. Korean J Urol. 2011;52:317–22.
Fritzsche FR, Oelrich B, Johannsen M, et al. Claudin-1 protein expression is a prognostic marker of patient survival in renal cell carcinomas. Clin Cancer Res. 2008;14:7035–42.
Tabaries S, Siegel PM. The role of claudins in cancer metastasis. Oncogene. 2017;36:1176–90.
Dudoit S, Yang YH, Callow MJ, et al. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat Sin. 2002;12:111–39.
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (2015R1A2A2A01006958) and the Asan Institute for Life Sciences, Asan Medical Center (2016-388).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Hwang, H.S., Go, H., Park, JM. et al. Epithelial-mesenchymal transition as a mechanism of resistance to tyrosine kinase inhibitors in clear cell renal cell carcinoma. Lab Invest 99, 659–670 (2019). https://doi.org/10.1038/s41374-019-0188-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-019-0188-y
This article is cited by
-
Protein tyrosine kinase inhibitor resistance in malignant tumors: molecular mechanisms and future perspective
Signal Transduction and Targeted Therapy (2022)
-
Resistance to tyrosine kinase inhibitors promotes renal cancer progression through MCPIP1 tumor-suppressor downregulation and c-Met activation
Cell Death & Disease (2022)
-
Identification of DNA methylation signatures associated with poor outcome in lower-risk Stage, Size, Grade and Necrosis (SSIGN) score clear cell renal cell cancer
Clinical Epigenetics (2021)
-
Vessel co-option and resistance to anti-angiogenic therapy
Angiogenesis (2020)