Abstract
Ki67 expression has been a valuable prognostic variable in breast cancer, but has not seen broad adoption due to lack of standardization between institutions. Automation could represent a solution. Here we investigate the reproducibility of Ki67 measurement between three image analysis platforms with supervised classifiers performed by the same operator, by multiple operators, and finally we compare their accuracy in prognostic potential. Two breast cancer patient cohorts were used for this study. The standardization was done with the 30 cases of ER+ breast cancer that were used in phase 3 of International Ki67 in Breast Cancer Working Group initiatives where blocks were centrally cut and stained for Ki67. The outcome cohort was from 149 breast cancer cases from the Yale Pathology archives. A tissue microarray was built from representative tissue blocks with median follow-up of 120 months. The Mib-1 antibody (Dako) was used to detect Ki67 (dilution 1:100). HALO (IndicaLab), QuantCenter (3DHistech), and QuPath (open source software) digital image analysis (DIA) platforms were used to evaluate Ki67 expression. Intraclass correlation coefficient (ICC) was used to measure reproducibility. Between-DIA platform reproducibility was excellent (ICC: 0.933, CI: 0.879–0.966). Excellent reproducibility was found between all DIA platforms and the reference standard Ki67 values of Spectrum Webscope (QuPath-Spectrum Webscope ICC: 0.970, CI: 0.936–0.986; HALO-Spectrum Webscope ICC: 0.968, CI: 0.933–0.985; QuantCenter-Spectrum Webscope ICC: 0.964, CI: 0.919–0.983). All platforms showed excellent intra-DIA reproducibility (QuPath ICC: 0.992, CI: 0.986–0.996; HALO ICC: 0.972, CI: 0.924–0.988; QuantCenter ICC: 0.978, CI: 0.932–0.991). Comparing each DIA against outcome, the hazard ratios were similar. The inter-operator reproducibility was particularly high (ICC: 0.962–0.995). Our results showed outstanding reproducibility both within and between-DIA platforms, including one freely available DIA platform (QuPath). We also found the platforms essentially indistinguishable with respect to prediction of breast cancer patient outcome. Results justify multi-institutional DIA studies to assess clinical utility.
Similar content being viewed by others

Introduction
Ki67 labeling index (Ki67 LI) is currently one of the most promising yet controversial biomarkers in breast cancer [1]. The European Society for Medical Oncology (ESMO) Clinical Practice Guidelines suggests that Ki67 LI may provide useful information, if the assay can be standardized [2]. The St. Gallen Consensus Conference in 2017 also agreed that Ki67 LI could be used to distinguish between HER2-negative luminal A-like and luminal B-like breast cancer subtypes [3]. However, the panel also emphasized the reproducibility issue of Ki67 LI, suggesting calibration of Ki67 scoring [3]. The American Society of Clinical Oncology recommended against the use of Ki67 LI for prognosis in newly diagnosed breast cancer patients because of lack of reproducibility across laboratories [4]. The International Ki67 in Breast Cancer Working Group (IKWG) has nevertheless published consensus recommendations for the application of Ki67 IHC in daily practice [5]. According to this group, parameters that predominantly influence the Ki67 IHC results include pre-analytical (type of biopsy and tissue handling), analytical (IHC protocol), interpretation and scoring, and data analysis steps [5]. Although these IKWG recommendations provide a guideline to improve pre-analytical and analytical consistency, inter-laboratory protocols still showed poor reproducibility related to different sampling, fixation, antigen retrieval, staining, and scoring methods [5, 6]. As the latter was the largest single contributor to assay variability, the IKWG has undertaken efforts to standardize visual scoring of Ki67 [7, 8], which requires on-line calibration tools and careful scoring of several hundred cells, which may or may not be practical for surgical pathologists to implement.
The emergence of digital image analysis (DIA) platforms has improved capacity and automation in biomarker evaluation [9]. DIA platforms are able to assess Ki67 LI and several studies have been conducted to compare manual scoring with DIA platforms [10,11,12,13,14]. However, different platforms have unique algorithms to segment and classify tissue and cellular compartments [10,11,12, 14, 15]. Finally, comparison studies of different platforms have been done by the International Ki67 Working Group, but not yet published at the time of this submission.
In this study, reproducibility of Ki67 LI was investigated across three DIA platforms with supervised classifiers applied by the same operator and by multiple operators. The effect of different training methods on automated Ki67 scoring was also investigated. In addition, by applying to an annotated series of breast cancers, the outcome prediction potential of the DIA platforms was also compared to each other.
Materials and methods
Patients
Two distinct breast cancer patient cohorts were employed in these investigations, totaling 179 patients. Cohort 1 is represented by 30 cases of ER+ breast cancer that were used in phase 3 of IKWG initiatives [7]. No survival data was available for this cohort, which was approved for the study by the British Columbia Cancer Agency’s Clinical Research Ethics Board (H10-03420). Cohort 2 comprises 149 breast cancer cases from the Pathology Department, Yale University, New Haven, CT, USA diagnosed between 1976 and 2003, with 120 months median follow-up for breast cancer-specific survival and 112 months median follow-up for relapse-free survival (RFS). The patient age at diagnosis ranged from 31 to 85 years, with a median age at diagnosis of 56 years (Table 1). All patients’ breast cancers had been surgically excised. Pathological features were retrieved from the pathology reports or the original H&E-stained slides were reviewed. This patient cohort was approved for the study by the Yale Human Investigation Committee under protocol #9505008219.
Tissue preparation and immunohistochemistry (IHC)
Preparation of the Ki67 slides of the first cohort has been previously described [7]. Briefly, the 30 core-cut biopsy blocks were centrally cut and stained with Ki67, resulting in 30 Ki67 slides from 30 cases. The IHC reaction was performed using monoclonal antibody Mib-1 at dilution 1:50 (DAKO UK, Cambridgeshire, UK) using an automated staining system (Ventana Medical Systems, Tucson, AZ, USA) according to the consensus criteria established by the International Ki67 Working Group [5].
In the second cohort, a tissue microarray was built from representative 10% neutrally buffered FFPE tissue blocks. Tumor areas were selected by pathologists based on hematoxylin and eosin-stained slides. Duplicate cores (each 0.6 mm in diameter) were punched from each case. The Mib-1 mouse monoclonal antibody (Dako, Carpinteria, CA, USA) was used to detect Ki67 [16]. This antibody had been previously validated by our research group [17, 18]. Slides were deparaffinized by heating for 1 h at 60 °C and soaked in xylene twice for 20 min, and were rehydrated in ethanol (twice in 100% ethanol for 1 min, twice in 95% ethanol for 1 min, once in 85% ethanol, and once in 75% ethanol). Antigen retrieval was performed in a PT module (LabVision, Fremont, CA, USA) with citrate buffer (pH 6.0) at 97 °C for 20 min. Endogenous peroxidase activity was blocked with hydrogen peroxide in methanol at room temperature for 30 min. Non-specific antigens were blocked with incubation in 0.3% bovine serum albumin in Tris-buffered saline/Tween for 30 min. Slides were then incubated with Ki67 mouse monoclonal antibody (1:100 dilution) for 1 h at room temperature. Next, slides were incubated in mouse EnVision reagent (Dako) for 1 h at room temperature. The EnVision reagent contains a mouse secondary antibody conjugated to many molecules of horseradish peroxidase (HRP). Slides were then incubated in hematoxylin and DAB to detect reactions.
Digital image analysis (DIA)
The Aperio ScanScope XT platform was used at ×40 to digitize the slides. Three different DIA platforms were used to evaluate Ki67 LI as follows: HALO (IndicaLab, Corrales, NM, USA), QuantCenter (3DHistech, Budapest, Hungary), and QuPath (open source software [19]). All software use color deconvolution, cell segmentation algorithms (e.g., Watershed cell detection) and supervised classifiers as machine-learning methods [20,21,22]. All these DIA platforms were trained to identify tumor cells, stromal cells, and immune cells (Figs. 1 and 2). As a ground truth to the machine-learning methods, we also evaluated Ki67 LI with meticulous manual tissue segmentation (crop areas of stromal and immune compartments) after automatic cell segmentation using the Spectrum Webscope platform with Nuclear algorithm (Aperio). When the reproducibility of Ki67 LI and the effect of different training methods on the Ki67 LI were investigated among three DIA platforms, all DIA were performed by an MD post-doc with expertise in breast pathology. As a machine-learning method, we used a random forest supervised classifier in QuPath and HALO platforms [23]; while QuantCenter applies wavelet-based multilevel feature extraction for pattern recognition [24, 25]. After setting the optimal color deconvolution and cell segmentation in all DIA platforms, training of the machine-learning methods was performed on the core-cut biopsy slides (first cohort) as follows: (I) Training was performed on one randomly selected slide (DIA 1). (II) Repeat training on the same slide at least 4 days later (DIA 1.1). (III) Training on another randomly selected slide (DIA 1.2). (IV) Training on five randomly selected slides (DIA 5). (V) Repeat training on the same five slides at least 4 days later (DIA 5.1). (VI) Training on another randomly selected five slides (DIA 5.2). For the QuantCenter software, the training was only possible on one slide. Thus, training methods IV–VI were not applied for QuantCenter. When outcome prediction potential of the DIA platforms was investigated, the same color deconvolution and cell segmentation settings were applied. The training of the machine-learning method was performed on one slide, because the second cohort consisted of one tissue microarray block.
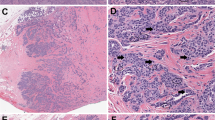
Representative pictures of digital image analysis (DIA) masks on a low cellular density breast cancer case (a). The first step of analysis with HALO (b) and QuantCenter (d) is the training of machine-learning classification to identify a tissue pattern (in this case areas of tumor cells) to be scored. Then, the cell segmentation is only applied in the annotations designated by the machine-learning classification. Thus, only tumor cells are shown in the DIA masks for both HALO and QuantCenter. Blue indicates negative tumor cells, and yellow, orange, and red indicate 1+, 2+, and 3+ positive tumor cells. In QuPath (c), the order of operations is switched, so that cell segmentation is the first, followed by machine-learning classification to identify a sub-population of cells to be scored (in this case tumor cells). Green indicates stromal cells, purple marks immune cells, blue corresponds to negative tumor cells, and yellow, orange, and red indicate 1+, 2+, and 3+ positive tumor cells
Representative pictures of digital image analysis (DIA) masks on a high cellular density breast cancer case (a). The first step of analysis with HALO (b) and QuantCenter (d) is the training of machine-learning classification to identify the tissue pattern (in this case areas of tumor cells) to be scored. Then, the cell segmentation is only applied in the annotations designated by the machine-learning classification. Thus, only tumor cells are shown in the DIA masks for both HALO and QuantCenter. Blue indicates negative tumor cells, and yellow, orange, and red indicate 1+, 2+, and 3+ positive tumor cells. In QuPath (c), the order of operations is switched, so that cell segmentation is the first, followed by machine-learning classification to identify a sub-population of cells to be scored (in this case tumor cells). Green indicates stromal cells, purple marks immune cells, blue corresponds to negative tumor cells, and yellow, orange, and red indicate 1+, 2+, and 3+ positive tumor cells
When the reproducibility of Ki67 LI was investigated among four different operators (A–D), QuPath was used to evaluate Ki67 LI. One of the four operators was an experienced breast pathologist, two of the four were MD post-docs with expertise in breast pathology. One operator out of the four was a post-doc researcher with expertise in breast pathology. All the operators used the same color deconvolution and cell segmentation setting that has been previously optimized. However, all the operators were free to train the machine-learning method on the same slide by annotating cells into the following classes: tumor cells, stromal cells, and immune cells. At least 4 days later, the DIA training was repeated on the same slide. To minimize the effect of intra-platform variability to inter-operator reliability, the mean Ki67 LI value of the two DIA evaluations were compared among the operators.
Statistical analysis
For statistical analysis SPSS 22 software (IBM, Armonk, USA) was used. The reproducibility among DIA platforms and operators was estimated by calculating an intraclass correlation coefficient (ICC). We considered ICC value between 0.4 and 0.6 as moderate reliability, values between 0.61 and 0.8 indicate good reliability, and values greater than 0.8 indicate excellent reliability [26]. Kaplan–Meier analysis supported with log-rank test was executed to assess prognostic potential. Breast cancer-specific survival was defined as the elapsed time from the date of primary diagnosis of the tumor to the date of death caused by breast cancer, or when patients were last censored if died of non-breast cancer cause or still alive. RFS was defined as time from the date of primary diagnosis to the occurrence of first relapse. Data were visualized using boxplots spaghetti plots and heat maps.
Results
Reproducibility of Ki67 LI among DIA platforms and training methods
Reproducibility among the DIA platforms was excellent (ICC: 0.933, CI: 0.879–0.966). The between-DIA platform reproducibility was better when applying training on five slides (ICC: 0.974, CI: 0.944–0.988) compared to applying training on one slide (ICC: 0.921, CI: 0.836–0.962, Fig. 3). QuPath returned systematically lower Ki67 LI results compared to HALO and QuantCenter platforms (Fig. 4). All DIA platforms showed excellent reproducibility with the reference standard Ki67 LI values of Spectrum Webscope (QuPath-Spectrum Webscope ICC: 0.970, CI: 0.936–0.986; HALO-Spectrum Webscope ICC: 0.968, CI: 0.933–0.985; QuantCenter-Spectrum Webscope ICC: 0.964, CI: 0.919–0.983).
Comparison of digital image analysis (DIA) platforms and different training methods. In spaghetti plots (a, b), each line represents Ki67 LI scores from one DIA platform, with a specific training method across the 30 cases. The bold black lines show Ki67 scores at 10, 20, and 30%. On the heat map of Ki67 scores (c), each row represents a case and each column represents a DIA platform, with a specific training method. Cases are ordered by the median scores (across DIA platforms)
Inter-platform variability of Ki67 LI. Since QuantCenter allowed to train only on one slide, a represents HALO and QuPath with one slide and five slides training. b shows all the platforms with only one slide training. The bottom/top of the box in each box plot represent the first (Q1)/third (Q3) quartiles, the bold line inside the box represents the median and the two bars outside the box represent the lowest/highest datum still within 1.5× the interquartile range (Q3–Q1). Outliers are represented with circles and extreme outliers with asterisk
The intra-DIA reproducibility was also excellent for all platforms (QuPath ICC: 0.992, CI: 0.986–0.996; HALO ICC: 0.972, CI: 0.924–0.988; QuantCenter ICC: 0.978, CI: 0.932–0.991). QuPath had the highest intra-DIA platform reproducibility and the lowest variability (Fig. 5). QuPath did not show systematic change in Ki67 LI values whether the DIA training was performed on one slide or five slides. However, HALO Ki67 LI values were systematically lower when the DIA training was performed on five slides compared to training performed on one slide (Fig. 3).
Prognostic potential of DIA platforms regarding Ki67 LI
For assessing breast cancer prognosis by Ki67 LI using a 10% cutpoint, either QuPath, or HALO and QuantCenter could perform statistically significant splitting of our cohort into patients’ group with statistically significant breast cancer-specific survival or RFS differences (Fig. 6). The hazard ratios of the DIA platforms ranged from 2.7 to 3.7, but were all comparable, with broadly overlapping 95% confidence intervals.
Reproducibility among four operators using the same DIA platform to evaluate Ki67 LI
The between-DIA platform reproducibility was excellent among all DIA platforms and QuPath showed the lowest intra-DIA platform variability. Thus, we selected the QuPath platform to investigate the reproducibility of Ki67 LI across four different operators using DIA. The four operators using QuPath to evaluate Ki67 LI had excellent reproducibility (ICC: 0.990, CI: 0.959–0.996, Fig. 7). The intra-rater reproducibility was also excellent for all the operators. The lowest intra-rater reproducibility was 0.962 (CI: 0.799–0.990), and the highest 0.995 (CI: 0.962–0.998).
Comparison of four operators (a–d) using the same DIA platform. In spaghetti plots (a–e), each line represents Ki67 LI scores from one operator across the 30 cases. The bold black lines show Ki67 scores at 10, 20, and 30%. a shows interobserver variability based on the mean of two evaluations for each operator. b–e represent the intra-observer variability for each operator based on two evaluations (1 and 1.1). On the heat map of Ki67 scores (f), each row represents a case and each column represents an operator. Cases are ordered by the median scores (across operators)
Discussion
Although it has long been acknowledged that detection of Ki67-positive tumor cells might provide prognostic and predictive information in breast cancer [27,28,29,30], it has not been widely adopted for clinical breast cancer management due to inter-operator and inter-institutional variability. Key contributors to variability include pre-analytical and technical aspects, and most significantly, lack of reproducibility in scoring across laboratories [6, 8]. Adding DIA to the pathological evaluation could improve standardization of this aspect of Ki67 assessment.
As the algorithms of several DIA platforms process staining patterns differently, cellular features, tissue morphology, and inter-DIA platform variability affect the Ki67 score [12, 31]. Thus, we aimed to investigate the inter-platform reproducibility across three independent DIA platforms applied to the same scanned images. Our results showed an excellent reproducibility (ICC: 0.933) among different DIA platforms suggesting that Ki67 LI could be broadly adopted if the DIA platform used at a given institution was calibrated to some centralized standard. To the best of our knowledge, no similar study has yet been published, wherein inter-platform reproducibility was investigated in Ki67 LI scoring between DIA platforms using different machine-learning methods. In a recent study by Koopman et al., the inter-platform agreement was investigated in Ki67 LI between two DIA platforms using virtual dual staining (VSD) [32]. The authors stained adjacent sections for cytokeratin (CK) 8/18 and Ki67. Then, the corresponding sections were digitally aligned to score Ki67 LI in the CK-positive areas. These authors found a very high correlation between two DIA platform using VSD. However, neither machine-learning methods to distinguish tissue patterns nor prognostic potential of DIA platforms regarding Ki67 LI was investigated [32]. In another detailed study by Paulik et al., the authors developed a DIA algorithm to detect cell nuclei in different IHC and FISH-stained breast samples [33]. The authors compared the sensitivity and positive predictive values (PPV) of their own and other DIA platforms in cell nuclei detection using manual nuclear marking as a reference standard. Although the authors revealed that the DIA platforms had PPV values in a range between 87 and 94%, the inter-platform reproducibility in Ki67 LI was not investigated [33].
Calibration and validation is crucial to the success of DIA [34]. In our study, all DIA platforms achieved excellent reproducibility with the reference standard (ICC: 0.964–0.970). This observation suggests that standardization of the platforms may be required for highly reproducible scores in Ki67 evaluation. Therefore, we investigated the effect of different training methods on automated Ki67 scoring. Our results revealed that the inter-platform reproducibility was better when applying training on five slides compared to applying training on one slide. The intra-platform reproducibility was also excellent in all investigated DIA platforms (ICC: 0.972–0.992). Operators can also affect outcome when using DIA. We found an ICC of 0.990 (CI: 0.959–0.996) among four operators using the same, calibrated DIA platform suggesting a highly reproducible Ki67 scoring method. Finally, it is always best to measure a new approach against outcome, rather than a previous method (in this case pathologist-read Ki67). Using annotated retrospective cohorts, our study showed that the automated Ki67 LI score of all three investigated DIA platforms was suitable to separate patients into good and unfavorable prognosis groups.
There are a number of limitations in this study. One limitation is study size. We used a low number of operators using DIA platform to evaluate Ki67, which may affect the power of the inter-rater results. We were also only able to test three software packages. Further studies are needed to investigate whether the training of different DIA platforms with machine-learning algorithms affects inter-laboratory reproducibility, especially in case of staining protocol differences or using different slide scanners.
In conclusion, our results suggest that DIA can be fairly easily standardized and may lead to highly reproducible, platform-independent scores in Ki67 evaluation. Our results suggest that automated Ki67 scoring could be independent of platform, operator, or vendor. We believe that this study is the first step of the standardization of automated DIA systems and also a step toward to utilizing Ki67 LI in clinical care. A multi-institutional DIA study is underway to prove clinical validity and utility.
References
Kos Z, Dabbs DJ. Biomarker assessment and molecular testing for prognostication in breast cancer. Histopathology. 2016;68:70–85.
Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E, et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(Suppl 5):v8–30.
Curigliano G, Burstein HJ, P Winer E, Gnant M, Dubsky P, Loibl S, et al. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol. 2017;28:1700–12.
Harris LN, Ismaila N, McShane LM, Andre F, Collyar DE, Gonzalez-Angulo AM, et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2016;34:1134–50.
Dowsett M, Nielsen TO, A′Hern R, Bartlett J, Coombes RC, Cuzick J, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103:1656–64.
Polley MY, Leung SC, McShane LM, Gao D, Hugh JC, Mastropasqua MG, et al. An international Ki67 reproducibility study. J Natl Cancer Inst. 2013;105:1897–906.
Leung SCY, Nielsen TO, Zabaglo L, Arun I, Badve SS, Bane AL, et al. Analytical validation of a standardized scoring protocol for Ki67: phase 3 of an international multicenter collaboration. NPJ Breast Cancer. 2016;2:16014.
Polley MY, Leung SC, Gao D, Mastropasqua MG, Zabaglo LA, Bartlett JM, et al. An international study to increase concordance in Ki67 scoring. Mod Pathol. 2015;28:778–86.
Kayser K, Gortler J, Borkenfeld S, Kayser G. How to measure diagnosis-associated information in virtual slides. Diagn Pathol. 2011;6(Suppl 1):S9.
Zhong F, Bi R, Yu B, Yang F, Yang W, Shui R. A comparison of visual assessment and automated digital image analysis of Ki67 labeling index in breast cancer. PLoS ONE. 2016;11:e0150505.
Klauschen F, Wienert S, Schmitt WD, Loibl S, Gerber B, Blohmer JU, et al. Standardized Ki67 diagnostics using automated scoring-clinical validation in the GeparTrio Breast Cancer Study. Clin Cancer Res. 2015;21:3651–7.
Stalhammar G, Fuentes Martinez N, Lippert M, Tobin NP, Molholm I, Kis L, et al. Digital image analysis outperforms manual biomarker assessment in breast cancer. Mod Pathol. 2016;29:318–29.
Stalhammar G, Robertson S, Wedlund L, Lippert M, Rantalainen M, Bergh J, et al. Digital image analysis of Ki67 in hot spots is superior to both manual Ki67 and mitotic counts in breast cancer. Histopathology. 2017;72:974–989.
Acs B, Madaras L, Kovacs KA, Micsik T, Tokes AM, Gyorffy B, et al. Reproducibility and prognostic potential of Ki-67 proliferation index when comparing digital-image analysis with standard semi-quantitative evaluation in breast cancer. Pathol Oncol Res. 2018;24:115–27.
Roge R, Riber-Hansen R, Nielsen S, Vyberg M. Proliferation assessment in breast carcinomas using digital image analysis based on virtual Ki67/cytokeratin double staining. Breast Cancer Res Treat. 2016;158:11–19.
Cattoretti G, Becker MH, Key G, Duchrow M, Schluter C, Galle J, et al. Monoclonal antibodies against recombinant parts of the Ki-67 antigen (MIB 1 and MIB 3) detect proliferating cells in microwave-processed formalin-fixed paraffin sections. J Pathol. 1992;168:357–63.
Bordeaux J, Welsh A, Agarwal S, Killiam E, Baquero M, Hanna J, et al. Antibody validation. Biotechniques. 2010;48:197–209.
Neumeister VM, Anagnostou V, Siddiqui S, England AM, Zarrella ER, Vassilakopoulou M, et al. Quantitative assessment of effect of preanalytic cold ischemic time on protein expression in breast cancer tissues. J Natl Cancer Inst. 2012;104:1815–24.
Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878.
Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23:291–9.
Fernandez R, Das P, Mirabet V, Moscardi E, Traas J, Verdeil JL, et al. Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nat Methods. 2010;7:547–53.
Amancio DR, Comin CH, Casanova D, Travieso G, Bruno OM, Rodrigues FA, et al. A systematic comparison of supervised classifiers. PLoS ONE. 2014;9:e94137.
Breiman L. Random forests. Mach Learn. 2001;45:5–32.
Scheunders P, Livens S, Van de Wouwer G, Vautrot P, Van Dyck D. Wavelet-based texture analysis. Int J Comput Sci Inf Manag. 1998;1:22–34.
Pittner S, Kamarthi SV. Feature extraction from wavelet coefficients for pattern recognition tasks. IEEE Trans Pattern Anal Mach Intell. 1999;21:83–88.
Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–74.
Stuart-Harris R, Caldas C, Pinder SE, Pharoah P. Proliferation markers and survival in early breast cancer: a systematic review and meta-analysis of 85 studies in 32,825 patients. Breast. 2008;17:323–34.
Criscitiello C, Disalvatore D, De Laurentiis M, Gelao L, Fumagalli L, Locatelli M, et al. High Ki-67 score is indicative of a greater benefit from adjuvant chemotherapy when added to endocrine therapy in luminal B HER2 negative and node-positive breast cancer. Breast. 2014;23:69–75.
Brown JR, DiGiovanna MP, Killelea B, Lannin DR, Rimm DL. Quantitative assessment Ki-67 score for prediction of response to neoadjuvant chemotherapy in breast cancer. Lab Invest. 2014;94:98–106.
Acs B, Zambo V, Vizkeleti L, Szasz AM, Madaras L, Szentmartoni G, et al. Ki-67 as a controversial predictive and prognostic marker in breast cancer patients treated with neoadjuvant chemotherapy. Diagn Pathol. 2017;12:20.
Kårsnäs A, Strand R, Doré J, Ebstrup T, Lippert M, Bjerrum K. A histopathological tool for quantification of biomarkers with sub-cellular resolution. Comput Methods Biomech Biomed Eng. 2015;3:25–46.
Koopman T, Buikema HJ, Hollema H, de Bock GH, van der Vegt B. Digital image analysis of Ki67 proliferation index in breast cancer using virtual dual staining on whole tissue sections: clinical validation and inter-platform agreement. Breast Cancer Res Treat. 2018;169:33–42.
Paulik R, Micsik T, Kiszler G, Kaszal P, Szekely J, Paulik N, et al. An optimized image analysis algorithm for detecting nuclear signals in digital whole slides for histopathology. Cytom A. 2017;91:595–608.
Laurinavicius A, Plancoulaine B, Laurinaviciene A, Herlin P, Meskauskas R, Baltrusaityte I, et al. A methodology to ensure and improve accuracy of Ki67 labelling index estimation by automated digital image analysis in breast cancer tissue. Breast Cancer Res. 2014;16:R35.
Acknowledgements
BA is supported by The Fulbright Program and The Rosztoczy Foundation Scholarship Program. DLR is supported by the Breast Cancer Research Foundation and grants from the NIH and his lab received instrument support from PerkinElmer and software support from Indica Labs and 3DHistech for this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
In the last 12 months, DLR has served as a consultant for advisor to Astra Zeneca, Agendia, Agilent, Biocept, BMS, Cell Signaling Technology, Cepheid, Merck, PerkinElmer, and Ultivue. TON reports a proprietary interest in PAM50/Prosigna and consultant work with NanoString Technologies. The remaining authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Acs, B., Pelekanou, V., Bai, Y. et al. Ki67 reproducibility using digital image analysis: an inter-platform and inter-operator study. Lab Invest 99, 107–117 (2019). https://doi.org/10.1038/s41374-018-0123-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-018-0123-7
This article is cited by
-
Clinical application of the HM-1000 image processing for HER2 fluorescence in situ hybridization signal quantification in breast cancer
Diagnostic Pathology (2024)
-
Subspecialized breast pathologists have suboptimal interobserver agreement in Ki-67 evaluation using 20% as the cutoff
Breast Cancer Research and Treatment (2024)
-
Endometrial cancer PDX-derived organoids (PDXOs) and PDXs with FGFR2c isoform expression are sensitive to FGFR inhibition
npj Precision Oncology (2023)
-
A Multi-Stain Breast Cancer Histological Whole-Slide-Image Data Set from Routine Diagnostics
Scientific Data (2023)
-
Exploration of Digital Image Analysis for Ki67 Quantification in the Grading of Medullary Thyroid Carcinoma: A Pilot Study with 85 Cases
Head and Neck Pathology (2023)