Abstract
Rho guanine nucleotide exchange factor 39 (ARHGEF39), also called C9orf100, is a new member of the Dbl-family of guanine nucleotide exchange factors. Although ARHGEF39 has been proven to regulate tumor progression in hepatocellular carcinoma, the downstream signaling pathway of ARHGEF39 and its clinical associations in non-small cell lung cancer (NSCLC) are currently unknown. In the present study, using MTT, colony formation, flow cytometry, mice xenografts, wound healing, and transwell assays, we showed that ARHGEF39 promoted tumor proliferation, migration, and invasion. Furthermore, ARHGEF39 promoted the expression of Cyclin A2, Cyclin D1, and MMP2 by activating Rac1, leading to increased phosphorylation of P38 and ATF2. Treatment with a P38 inhibitor counteracted the effect of ARHGEF39 overexpression on the increase in Cyclin A2, Cyclin D1, and MMP2 expression. Moreover, the elevated levels of p-P38 and p-ATF2 caused by ARHGEF39 overexpression could be inhibited by expression of a dominant negative Rac1 mutant (T17N). In addition, the inhibition of the expression of p-P38 and p-ATF2 by ARHGEF39 RNAi could be restored by the expression of a constitutively active Rac1 mutant (Q61L). A similar impact on cell growth and invasion was observed after ARHGEF39 overexpression combined with the P38 inhibitor, Rac1 T17N, or Rac1 Q61L. Using immunohistochemistry, ARHGEF39 expression was observed to correlate positively with larger tumor size in clinical samples from 109 cases of NSCLC (P = 0.008). The Kaplan–Meier test revealed that ARHGEF39 expression significantly affected the overall survival of patients with NSCLC (52.55 ± 6.40 months vs. 64.30 ± 5.40 months, P = 0.017). In conclusion, we identified that ARHGEF39 promotes tumor growth and invasion by activating the Rac1-P38-ATF2 signaling pathway, as well as increasing the expression of Cyclin A2, Cyclin D1, and MMP2 in NSCLC cells. ARHGEF39 may be a useful marker to predict poor prognosis of patients with NSCLC.
Similar content being viewed by others
Introduction
Rho guanine nucleotide exchange factor 39 (ARHGEF39), also called C9orf100, is a new member of the Dbl-family of guanine nucleotide exchange factors (GEFs) [1]. GEFs are known as important activators of Rho GTPases, which are significantly associated with cell migration [2,3,4,5]. The Dbl family is characterized by the presence of a Dbl homology (DH) catalytic domain, followed by an adjacent pleckstrin homology (PH) domain, C-terminal to the DH domain [6]. In most GEFs, the DH–PH domains are flanked by a diverse array of protein-protein and protein-lipid interaction domains [5, 6]. First, this diversity in flanking non-RhoGEF sequences distinguishes the regulatory roles of RhoGEFs that otherwise activate the same set of Rho GTPases. Second, these flanking domains may also help to regulate their intracellular localization and association with other proteins. Finally, these flanking sequences might facilitate scaffolding functions of RhoGEFs, which could further influence which effectors are activated [5]. In contrast to other family members, ARHGEF39 lacks the additional functional domains in its structure. Interestingly, besides control of migration, ARHGEF39 also promotes proliferation of human hepatocellular carcinoma cells [1, 5, 6]. However, little is known about the involvement in tumors of Dbl-family GEFs that have minimal flanking domains except for the DH and PH domains. Thus, the downstream signaling pathways of ARHGEF39 and its clinicopathological significance need to be determined.
The purpose of this study was to investigate the potential downstream signaling pathway of ARHGEF39 associated with regulation of the proliferation in non-small cell lung cancer (NSCLC) and to analyze the association between ARHGEF39 and clinicopathological features of patients with lung cancer.
Materials and methods
Patients and specimens
This study was conducted with the approval of the local institutional review board at the China Medical University (LS2016 [009]). Written consent was given by the participants for their information to be stored in the hospital database and for their specimens to be used in this study. Tissue samples were obtained from 109 patients (68 males and 41 females) who underwent complete surgical excision at the First Affiliated Hospital of China Medical University between 2010 and 2012, with a diagnosis of lung squamous cell carcinoma or lung adenocarcinoma. The median age of the 109 patients was 60 years old (ranging from 29 to 79 years old). Among the 109 patients, 49 were 60 years old or older, and 60 patients were less than 60 years old. The samples included 47 squamous cell lung carcinomas and 62 lung adenocarcinomas. A total of 38 tumors were well differentiated, while 71 were classified as moderately or poorly differentiated. Lymph node metastases were present in 48 of the 109 cases. The tumors included 83 at stages I–II and 26 at stage III. No neoadjuvant radiotherapy or chemotherapy was administered before surgery. Of the 109 patients, 33 (30.3%) were treated with platinum-based adjuvant chemotherapy, eight (7.3%) underwent platinum-based adjuvant chemoradiotherapy, and the other 68 patients were not treated in our hospital; thus, we did not have information about their treatment. The survival of each patient was defined as the time from the day of surgery to the end of follow-up or the day of death. Histological diagnosis and grading were evaluated according to the 2015 World Health Organization (WHO) classification of tumors of the lung [7]. All 109 specimens were examined for histological subtype, differentiation, and tumor stage. Tumor staging was performed according to the seventh edition of the Union for International Cancer Control (UICC) TNM Staging System for Lung Cancer [8]. Sixteen freshly isolated specimens, including both tumor tissue and the corresponding normal tissues, were stored at −80 °C immediately after resection for protein extraction.
Cell culture and transfection
The HBE cell line was obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The A549, H460, H661, H292, 83A, and H1299 cell lines were obtained from the Shanghai Cell Bank (Shanghai, China). The LK2 cell line was a gift from Dr. Hiroshi Kijima (Department of Pathology and Bioscience, Hirosaki University Graduate School of Medicine, Hirosaki, Japan). All cell lines were authenticated by short tandem repeat (STR) DNA profiling. Upon receipt, cells were frozen, and individual aliquots were cultured and were typically analyzed within 10 passages. All cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 100 IU/ml penicillin (Sigma, St. Louis, MO, USA), and 100 μg/ml streptomycin (Sigma), and passaged every other day using 0.25% trypsin (Invitrogen).
Plasmids pCMV6-ddk-myc and pCMV6-ddk-myc-ARHGEF39 were purchased from Origene (Rockville, MD, USA). Plasmids pRK5-myc-Rac1-T17N (Addgene, Cambridge, MA, USA #12984) and pRK5-myc-Rac1-Q61L (Addgene #12983) were kind gifts from Dr. Gary Bokoch Lab (Department of Immunology and Microbial Science and the Department of Cell Biology, The Scripps Research Institute, San Diego, CA, United States). ARHGEF39-siRNA (SR313727) and control NC-siRNA (SR30004) were purchased from Origene. Stable or transient transfection was carried out using the Lipofectamine 3000 reagent (Invitrogen), as previously described; each transfection was carried out in triplicate [9].
Western blotting analysis
Total protein was extracted using lysis buffer (Pierce, Rockford, IL, USA) and quantified using the Bradford method [10]. Fifty micrograms of the total protein samples were separated by 10% SDS-PAGE and transferred onto polyvinylidene fluoride membranes (PVDF; Millipore, Billerica, MA, USA). Membranes were incubated overnight at 4 °C with the following primary antibodies: anti-ARHGEF39 (1:100, HPA061299, Sigma); anti-GAPDH (WH0002597M1, 1:5000, Sigma); anti-Myc-tag (#2276), anti-Cyclin A2 (#4656), anti- Cyclin B1 (#4138), anti-Cyclin D1 (#2978), anti-Cyclin E1 (#4129), anti-Cyclin H (#2927), anti-MMP2 (#13132), MMP9 (#2270), anti-phosphorylated (p)-P38 (#4511), P38 (#2308), p-ATF2 (#5112), ATF2 (#9226), anti-active-β-catenin (#19807), anti-β-catenin (#2698), anti-p-JNK (#4668), anti-JNK (#9252), anti-p-NF-κB (#13346), anti-NF-κB (#8242), anti-p-AKT (#12694), anti-AKT (#2920), anti-p-ERK (#4370), anti-ERK (#4695), anti-RhoA (#2117), anti-RhoC (#3430), and anti-(#2466) (1:500; Cell Signaling Technology, Danvers, MA, USA); and anti-Rac1 (sc-24567, Santa Cruz Biotechnology, Santa Cruz, CA, USA). Membranes were washed and subsequently incubated with peroxidase-conjugated anti-mouse or anti-rabbit IgG (Santa Cruz Biotechnology) at 37 °C for 2 h. Bound proteins were visualized using electrochemiluminescence (Pierce) and detected with a bio-imaging system (DNR Bio-Imaging Systems, Jerusalem, Israel).
Immunofluorescent staining
The cells were fixed with 4% paraformaldehyde, blocked with 1% bovine serum albumin (BSA) and then incubated with the ARHGEF39 (1:100, HPA061299, Sigma) overnight at 4 °C, followed by incubation with tetramethylrhodamine isothiocyanate (TRITC)-conjugated secondary antibodies (1:200, Zhongshan Golden Bridge, Beijing, China) at 37 °C. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Epifluorescent microscopy was performed using a Radiance 2000 laser scanning confocal microscope (Carl Zeiss, Thornwood, NY, USA).
Colony formation assay
A549 cells were stably transfected with pCMV6 or pCMV6-ARHGEF39 plasmids; H460 cells were transfected with negative control or ARHGEF39-siRNA. Thereafter, cells were plated into three 6-cm cell culture dishes (1000 cells per dish for A549 and H460 cell lines) and incubated for 12 days. Plates were washed with phosphate-buffered saline (PBS) and stained with Giemsa. The number of colonies with more than 50 cells was counted. The colonies were counted manually under a microscope. Each experiment was carried out in triplicate.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Cells stably transfected with pCMV6 or pCMV6-ARHGEF39 plasmids, and transfected with negative control or ARHGEF39-siRNA, were plated in 96-well plates in medium containing 10% FBS at about 3000 cells per well. For quantification of cell viability, cultures were stained after 4 or 5 days using the MTT assay. Briefly, 20 μl of 5 mg/ml MTT solution was added to each well and incubated for 4 h at 37 °C, the medium was then removed from each well, and the resultant MTT formazan was solubilized in 150 μl of dimethyl sulfoxide (DMSO). The results were quantified spectrophotometrically at a wavelength of 490 nm. Each test carried out in triplicate.
Cell cycle analysis
Flow cytometry was used to analyze the cell cycle according to standard methods on a FACS Calibur cytometer (Becton-Dickinson Biosciences, San Jose, CA, USA). For DNA content analysis, A549 cells stably transfected with either empty plasmid or ARHGEF39 plasmid, or H460 cells transfected with ARHGEF39-specific siRNA or control siRNA, were harvested at different time points, fixed in cold 70% ethanol, washed, and rehydrated in PBS. DNA staining was performed by treating the cells with RNase A (10 mg/ml) for 30 min and with propidium iodide (10 µg/ml; Sigma) for 5 min.
Xenograft model
The animals used in this study were treated according to the National Institutes of Health Guide for the care and use of Laboratory Animals (NIH Publications No. 8023, revised 1978). Four-week-old female BALB/c nude mice were purchased from SLAC Laboratory Animal (Shanghai, China) and kept in a laminar flow cabinet under specific pathogen-free conditions for two weeks before use. Mice were randomized into different groups (n = 5 per group). A549 cells stably transfected with either empty plasmid or the ARHGEF39 plasmid were injected subcutaneously into the nude mice. Bidimensional tumor measurements were taken at the 12th day and the 36th day; after which the mice were sacrificed. All experimental procedures were approved by the Animal Ethics Committee of the China Medical University.
Wound healing assay
A scratch was made using a 200- µl pipette tip after the cells were grown to 80% confluence. Cells were then incubated in medium containing 5% FBS. The gap size was measured 24 or 48 h later.
Invasion assay
The invasive abilities of the cells were evaluated using a transwell assay (Millipore, Bedford, MA, USA). For the invasion assay, transwells were pre-coated with 35 µl of 3× diluted matrix matrigel (Bd Biosciences Pharmingen, San Diego, CA, USA) for 30 min. Cells (2 × 105) in serum-free culture medium were added to the upper chamber of the device, and the lower chamber was filled with 10% FBS culture medium. After the indicated hours of incubation, the remaining cells on the upper surface of the filter were carefully removed with a cotton swab. The filter was then fixed, stained, and photographed. Migrated or invaded cells were quantified by counting the cells in three random fields per filter.
Pull-down assay of activated Rho GTPases
The cellular levels of GTP-loaded RhoA, Rac1, and Cdc42 were determined using a RhoA/Rac1/Cdc42 Activation Assay Kit (Cytoskeleton, Denver, CO, USA), according to the manufacturer’s instructions. Briefly, cells were washed with PBS and lysed in Mg2+ Lysis Buffer (MLB) after transfecting for 48 h. The lysates were clarified by centrifugation for 5 min at 14000×g and incubated with Rhotekin Ras-binding domain (RBD)-agarose or p21 activated kinase 1 protein (PAK-1) Rac/Cdc42 (p21) binding domain (PBD)-agarose for 45 min at 4 °C with rotation. The beads were pelleted and washed with MLB three times, and bound proteins were eluted with 40 µl of SDS sample buffer. Samples were separated by SDS-PAGE, followed by western blotting with anti-RhoA, anti-Rac1, and anti-Cdc42 antibodies.
Immunohistochemistry
Samples were fixed in 10% neutral formalin, embedded in paraffin, and sliced into 4-μm thick sections. Immunostaining was performed by the streptavidin-peroxidase method. The sections were incubated with the following antibodies: Anti-ARHGEF39 (1:100; HPA061299, Sigma), p-P38 (#4631), anti-p-ATF2 (#9221), anti-Cyclin D1 (#2978), anti-MMP2 (#13132), and anti-Ki-67 (#9027) (1:400, Cell Signaling Technology) at 4 °C overnight, followed by biotinylated goat anti-rabbit IgG secondary antibody. After washing, the sections were incubated with horseradish peroxidase-conjugated streptavidin-biotin (Ultrasensitive; MaiXin, Fuzhou, China) and developed using 3,3-diaminobenzidine tetra-hydrochloride (MaiXin). Finally, samples were lightly counterstained with hematoxylin, dehydrated in alcohol, and mounted. Two investigators who were blinded to the clinical data scored the slides semi-quantitatively by evaluating the staining intensity and percentage of stained cells in representative areas. The staining intensity was scored as 0 (no signal), 1 (weak), 2 (moderate), or 3 (high). The percentage of cells stained was scored as 1 (1–25%), 2 (26–50%), 3 (51–75%), or 4 (76–100%). A final score of 0–12 was obtained by multiplying the intensity by the percentage scores. Tumors were regarded as positive for ARHGEF39 expression if their score was > 4. Tumor samples with scores between 1 and 4 were categorized as showing weak expression, whereas those with scores of 0 were considered to have no expression; both weak expression and no expression were defined as negative ARHGEF39 expression.
Online analysis of overall survival in patients with NSCLC
To evaluate the relationship between the presence of ARHGEF39 and patient clinical outcome, we used the KM Plotter Online Tool in the patients with NSCLC. This is a public database containing information from 1928 patients that permit the investigation of the association of genes with overall survival (OS) [11].
Statistical analysis
SPSS version 22.0 for Windows (SPSS, Chicago, IL, USA) was used for all analyses. Pearson’s Chi-square test was used to assess possible correlations between ARHGEF39 and clinicopathological factors. The Mann–Whitney U test was used for the image analysis of colony formation assay, MTT, cell cycle assay, xenograft assay, wound healing assay, and transwell assay. Kaplan–Meier survival analysis was carried out for the 109 NSCLC cases and compared using the log-rank test. A P < 0.05 was regarded as statistically significant.
Results
Overexpression of ARHGEF39 promoted cell proliferation in vitro and in vivo
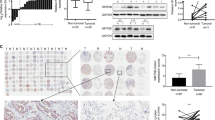
First, we evaluated the ARHGEF39 protein levels in the normal bronchial epithelial cell line (HBE) and in seven commonly used lung cancer cell lines using western blotting. As shown in Fig. 1a, positive expression of ARHGEF39 was observed in all eight cell lines. A subsequent immunofluorescence assay showed that ARHGEF39 was positively expressed in the cytoplasm of HBE, A549, and H460 cell lines; representative images are shown in Fig. 1b. To examine the association between ARHGEF39 and cell proliferation, we overexpressed ARHGEF39 in A549 cells. Western blotting analysis showed that ARHGEF39 was successfully overexpressed in the A549 cells (Fig. 1c). Cell viability assays revealed that cell transfected with the ARHGEF39 plasmid grew more rapidly than those transfected with the empty vector (Fig. 1d, P < 0.01). A colony formation assay was also performed to determine the long-term effects of ARHGEF39 on cell growth. We found that more colonies were formed in A549 cells transfected with ARHGEF39 than in the controls (Fig. 1e, f, P < 0.01). To understand the mechanism by which ARHGEF39 affects cell growth, we performed flow cytometry to investigate the cell cycle distribution of A549 cells at different time-points following ARHGEF39 plasmid transfection. As shown in Fig. 1g, h, overexpressing ARHGEF39 led to cell cycle arrest at S phase at 48 h. In line with the data from in vitro analyses, our in vivo experiments showed that the size and weight of the tumor xenografts were dramatically increased in mice injected with cells overexpressing ARHGEF39 compared those injected with cells transfected with the empty vector (Fig. 1i–k). After transfecting ARHGEF39 plasmids into A549 cells, the results of wound healing and transwell assays suggested that migration (Fig. 1l, m) and invasion (Fig. 1n, o) were obviously enhanced by the overexpression of ARHGEF39 (both P < 0.05).
High expression of ARHGEF39 (A39) promotes NSCLC proliferation, migration, and invasion in vitro and in vivo. a In six of seven NSCLC cell lines, the protein level of A39 was higher than that in HBE cells. b Representative images of immunofluorescent staining results for A39 in HBE, A549, and H460 cells. c Stable upregulation of A39 expression in the transfected A549 cell line was confirmed by western blotting. d–f Cell proliferation was significantly increased after A39 overexpression, as assessed by MTT and colony formation assays. g–h Cell-cycle analysis showing that A39 overexpression caused an S phase arrest. i–k The tumor size and weight of xenografts were measured at the indicated time points; in vivo tumor growth in the pCMV6-A39 group was significantly increased compared with that in the pCMV6-Vector group. l–m Wound healing migration assays and the quantification of the distance of the open area are shown. n–o The invasion of A549 cells was measured using transwell Matrigel invasion assays. Data are shown as the mean ±SD from three independent experiments. *P < 0.05, **P < 0.01; scale bar = 50 μm
Depletion of ARHGEF39 inhibited cell growth and induced cell cycle arrest
To further elucidate the function of ARHGEF39, we next examined whether a reduction of endogenous ARHGEF39 expression would suppress cell growth, migration, and invasion. After knocking down ARHGEF39 expression, western blotting was performed to evaluate the interference efficiency. The cells transfected with NC-siRNA were used as the control. Compared with the control, ARHGEF39-siRNA strongly decreased the protein level of endogenous ARHGEF39 in H460 cells (Fig. 2a). Depletion of ARHGEF39 significantly inhibited the cell growth (Fig. 2b) and colony formation ability of H460 cells (Fig. 2c, d). As shown in Fig. 2e, f, S phase arrest in H460 cells was reversed by downregulating endogenous ARHGEF39 expression at 48 h. Wound healing and transwell assays results showed that migration (Fig. 2g, h) and invasion (Fig. 2i, j) were obviously inhibited by knocking down ARHGEF39 (P < 0.05, respectively).
Knockdown of endogenous ARHGEF39 (A39) inhibited NSCLC proliferation, migration, and invasion in vitro. a Downregulation of A39 expression in the H460 cell line was confirmed by western blotting. b–d Cell proliferation was significantly decreased after A39 RNAi, as assessed by MTT and colony formation assays. e–f Cell-cycle analysis showed that A39 RNAi facilitated the transition from S phase to G1 phase. g–h Wound healing migration assays and the quantification of the distance of the open area are shown. i–j The invasion of H460 cells was measured using transwell Matrigel invasion assays. Data are shown as the mean ± SD from three independent experiments. *P < 0.05, **P < 0.01; scale bar = 50 μm
ARHGEF39 upregulated the expression of Cyclin A2 and Cyclin D1 via phosphorylation of the P38-ATF2 signaling pathway
Cyclins and MMPs play important roles in regulating tumor proliferation and invasion; therefore, we evaluated the protein levels of Cyclins and MMPs after overexpressing ARHGEF39 in A549 cells and depleting ARHGEF39 in H460 cells. The levels of Cyclin A2, Cyclin D1, and MMP2 were obviously increased after overexpressing ARHGEF39. Accordingly, their expression levels were downregulated when ARHGEF39 was depleted (Fig. 3a, b). The levels of Cyclin B1, Cyclin E1, Cyclin H, and MMP9 showed no visible changes (Supplementary Fig. 1). Using western blotting analysis, we also investigated multiple key proteins of the signaling pathways involved in regulating cell proliferation and the cell cycle. The levels of phosphorylated P38 and ATF2 were elevated after transfection with the ARHGEF39 cDNA, while they were decreased after transfection with the ARHGEF39 siRNA (Fig. 3a, b). There were no obvious changes of the level of P38, ATF2, Active-β-catenin, β-catenin, p-JNK, JNK, p-NF-κB, NF-κB, p-AKT, AKT, p-ERK, and ERK (Fig. 3a, b and Supplementary Fig. 1). Subsequently, SB203580, a P38 inhibitor, was incorporated into the medium of A549 cells with or without overexpression of ARHGEF39. Compared with the controls, although the levels of p-P38 and P38 were not visibly depressed, the expression of p-ATF2, a downstream molecule of P38 signaling, showed obviously downregulation. Interestingly, the elevated expression levels of Cyclin A2, Cyclin D1, and MMP2 induced by ARHGEF39 overexpression were prominently inhibited by SB203580 (Fig. 3c). Immunohistochemistry results showed that overexpression of ARHGEF39 led to upregulation of the expression levels of p-P38, p-ATF2, Cyclin D1, MMP2, and Ki-67 in nude mice xenografts (Fig. 3d). Subsequent colony formation assays and cell cycle analysis showed that the increasing number of clones and S phase arrest that were caused by transfection with the ARHGEF39 plasmid were counteracted by incorporation of SB203580 (Fig. 3e–h). A similar tendency was observed in the wound healing (Fig. 3i, k) and transwell assays (Fig. 3j, l).
ARHGEF39 (A39) promoted the expression of CyclinA2, Cyclin D1, and MMP2 by facilitating phosphorylation of the P38-ATF2 signaling pathway. a The levels of p-P38, p-ATF2, Cyclin A2, Cyclin D1, and MMP2 were elevated after transfection with A39 cDNA in A549 cells. (b) The levels of p-ATF2, Cyclin A2, Cyclin D1, and MMP2 were downregulated after A39 RNAi transfection in H460 cells. (c) The increasing levels of p-ATF2, Cyclin A2, Cyclin D1, and MMP2 caused by A39 overexpression were counteracted by incorporation of SB203580, a P38 phosphorylation inhibitor, into the growth media. d Representative images of the immunohistochemical staining results in mice xenografts; the expression levels of p-P38, p-ATF, Cyclin D1, MMP2, and Ki-67 were compared between A549-A39 cells and A549-vector cells. e–f In the colony formation assay, the increase in cell proliferation induced by A39 overexpression was reversed by adding SB203580. g–h Cell cycle analysis showing that SB203580 incorporation attenuated the effect of A39 overexpression on facilitating S phase arrest. i, k Wound healing and (j, l) Matrigel assays showing that inhibition of P38 phosphorylation using SB203580 abolished the increase in cancer cell migration and invasion induced by A39 overexpression. Data are shown as the mean ± SD from three independent experiments. *P < 0.05, **P < 0.01; scale bar = 50 μm
ARHGEF39 promoted the phosphorylation of the P38-ATF2 signaling pathway by activating Rac1
We examined whether the small GTPases Rac1, RhoA, RhoC, and Cdc42 might be activated by the overexpression of ARHGEF39. Following ARHGEF39 overexpression, active Rac1, RhoA, RhoC, and Cdc42 were immunoprecipitated. Only the activation of Rac1, but not RhoA, RhoC, and Cdc42, was increased (Fig. 4a). Subsequently, the Rac1 T17N plasmid (a dominant negative mutant of Rac1) was transfected into A549 cells, with or without ARHGEF39 overexpression. The increase in the level of GTP-Rac1 following overexpression of ARHGEF39 was counteracted by Rac1 T17N transfection, as were the increases in p-P38 and p-ATF2 expression. Similarly, inhibition of GTP-Rac1 caused by ARHGEF39 RNAi, as well as the inhibition of p-P38 and p-ATF2, was reversed by transfection with the Rac1 Q61L plasmid (a constitutively active mutant of Rac1; Fig. 4b). Subsequent MTT assays (Fig. 4c), colony formation assays (Fig. 4d, g), transwell analysis (Fig. 4e, h), and wound healing assay (Fig. 4f, i) showed similar tendencies to the western blotting results.
ARHGEF39 (A39) facilitated phosphorylation of P38-ATF2 by activating Rac1. a A549 cells transfected with empty vector or A39 cDNA were subjected to active Rac1, RhoA, RhoC, and Cdc42 pull-down assays. The amount of active or total Rac1, RhoA, RhoC, and Cdc42 are shown, respectively. b Transfection with a plasmid encoding Rac1 (T17N) attenuated the increase of active Rac1, as well as the levels of p-P38 and p-ATF2, induced by A39 overexpression. Transfection with a plasmid encoding Rac1 (Q61L) restored the decrease of active Rac1, as well as the levels of p-P38 and p-ATF2, caused by A39 RNAi. In MTT assays (c), colony formation assays (d, g), wound healing assays (f, i), and Matrigel assays (e, h), the increase in cell proliferation, migration, and invasion, induced by A39 overexpression, was abolished by transfection with Rac1 (T17N), whereas the inhibition of cell proliferation, migration and invasion, caused by A39 RNAi, was restored following transfection with Rac1 (Q61L). Data are shown as the mean ± SD from three independent experiments. *P < 0.05, **P < 0.01; scale bar = 50 μm
ARHGEF39 positively expressed in the cytoplasm and/or membrane of lung cancer cells, and predicted poor clinical outcomes of patients with NSCLC
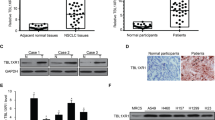
We evaluated the protein levels of ARHGEF39 in clinical NSCLC tissue samples using western blotting. ARHGEF39 was expressed at higher levels in the lung cancer tissues of 11/16 cases (68.8%), compared with the adjacent noncancerous tissues. Image analysis of the western blot demonstrated that the mean ± SE normalized ARHGEF39 protein expression level in NSCLC (0.89 ± 0.07) was significantly higher than that in the corresponding noncancerous tissues (0.50 ± 0.08, P = 0.0105; Fig. 5a, b).
Expression and prognostic values of ARHGEF39 (A39) in NSCLC tissues. Western blotting detection (a) and the quantification (b) of A39 expression in 16 paired tumors (T) and peritumoral normal lung tissues (N); β-actin served as a loading control. c Representative figures showing negative or weak expression of A39 in normal bronchial epitheliums (a), alveoli (b); and positive cytoplasmic (c) or cytoplasmic/membranous (d) expression of A39 in squamous cell carcinomas; and positive cytoplasmic (e) or cytoplasmic/membranous (f) expression of A39 in adenocarcinomas. d Kaplan–Meier survival analysis revealed that the overall survival of patients with positive A39 expression was significantly shorter than those with negative A39 expression. e Results of online software survival prediction (KM Plotter). *P < 0.05; scale bar = 50 μm
Subsequent immunohistochemical analysis showed that ARHGEF39 was positively expressed in the cytoplasm and/or membrane of lung cancer tissues samples (Fig. 4c). Of the 109 NSCLC tissue samples, ARHGEF39 was positively expressed in 62 cases (56.9%), and 25 of the 62 cases had membranous expression. Interestingly, only five cases with membranous expression showed no cytoplasmic expression, the other 20 cases presented both membranous and cytoplasmic ARHGEF39 expression.
As shown in Table 1, ARHGEF39 expression correlated positively with larger tumor size in the 109 cases of NSCLC (P = 0.008). There was no significant association between ARHGEF39 expression and age, gender, TNM staging, regional lymph node metastasis, histological differentiation, histological type, and smoking history. The Kaplan–Meier test revealed that ARHGEF39 expression significantly affected the OS of patients with NSCLC (Fig. 5d). The OS of patients with NSCLC with positive ARHGEF39 expression (52.55 ± 6.40 months) was significantly shorter than patients with negative ARHGEF39 expression (64.30 ± 5.40 months, P = 0.017). Furthermore, we also used the KM-plotter tool, as described in Material and Methods section, which contains information from datasets grouping 1928 NSCLC patients, to predict the impaction of ARHGEF39 gene expression on OS. As can be seen in Fig. 5e, we identified that the ARHGEF39 gene is associated with worse outcome in patients with NSCLC for OS (P < 0.001).
Discussion
The results of the present study revealed that ARHGEF39 enhanced tumor proliferation and invasion by activating its downstream Rac1-P38-ATF2 signaling pathway in NSCLC cells. Positive ARHGEF39 expression was associated with larger tumor size and was a potential predictor of poor prognosis of patients with NSCLC.
ARHGEF39 is a novel member of the Dbl-family of GEFs [1]. The main function of Dbl-family GEFs is to regulate the activation of Rho GTPases, which is critical to a variety of cellular events, such as cell cycle progression, cytoskeletal reorganization, membrane trafficking, cell migration, and invasion [2, 3, 12, 13]. As a member of Dbl RhoGEFs, ARHGEF39 shares conserved DH and PH domains with the other family members, which indicated that ARHGEF39 might retain the main functions of Dbl RhoGEFs. However, ARHGEF39 presents minimal N- and C-terminal sequences flanking the DH/PH domains compared with most other family members [1, 5, 6]. These flanking non-RhoGEF sequences determine the activation of different RhoGTPases and lead to their utilization of distinct effectors. The downstream signaling pathway and their effectors associated with ARHGEF39 are currently unknown. In a previous study, ARHGEF39 was found to promote cell proliferation by inducing cell cycle arrest in hepatocellular carcinoma cells, which suggested that ARHGEF39 functioned in regulating tumorigenesis and progression [1]. Our results were consistent with previous data that transfection of exogenous ARHGEF39 facilitated cell growth, migration, and invasion in vitro and in vivo. Similar results were also observed by knockdown of endogenous ARHGEF39 [1].
In the present study, for the first time, we identified that ARHGEF39 upregulated Cyclin A2, Cyclin D1, and MMP2 by activating the Rac1-P38-ATF2 signaling pathway, which might contribute to promoting cell proliferation and invasion. Rho GTPases had been shown to regulate P38 signaling through activating Rac1 [14, 15]. We also performed an immunoprecipitation assay to explore the interaction between ARHGEF39 and Rac1, but did not find a direct interaction. As a result, we postulated that the activation of Rac1 by ARHGEF39 may be indirect (data not shown). Certainly, the underlying mechanisms need further investigation. In previous studies, ARHGEF39 was predicted to interact with DVL2, which is an important regulator of Rho GTPases [16,17,18]. Therefore, we also examined the effect of ARHGEF39 on DVL2. Unfortunately, immunoprecipitation assays did not show any interaction between ARHGEF39 and DVL2 (data not shown). These results may have resulted from a number of factors, including the type of assay performed, types of transfected cells, conditions of transfection, or other reasons.
ARHGEF39 was found to be frequently upregulated in hepatocellular carcinoma samples and was associated with clinicopathological features of hepatocellular carcinoma patients [1]. However, the clinicopathological relevance of ARHGEF39 in NSCLC, especially the association with the clinical outcomes of lung cancer patients, was largely unknown until now. In the present study, ARHGEF39 was also frequently upregulated in NSCLC samples compared with corresponding noncancerous tissues. Positivity for ARHGEF39 correlated with larger tumor size and predicted poor prognosis of patients with NSCLC. These data strongly indicated that ARHGEF39 mediated the development and progression of NSCLC. Information on treatment after surgery was limited; therefore our study could not perform an analysis of progression-free survival. More studies are required to confirm the prognostic value of ARHGEF39. Another limitation of our study was that ARHGEF39 showed cytoplasmic and/or membranous expression in clinical tissue samples while it only presented cytoplasmic expression in the cell lines. The results of the experiment in cell lines could not indicate the function of the membranous ARHGEF39. This aspect also requires further experiments. Taken together, we identified that ARHGEF39 promotes tumor growth and invasion by activating the Rac1-P38-ATF2 signaling pathway and upregulating the protein levels of Cyclin A2, Cyclin D1, and MMP2 in NSCLC cells. ARHGEF39 is frequently upregulated in NSCLC tissue samples and is associated with lung cancer progression. ARHGEF39 may be a useful marker to predict poor prognosis of patients with NSCLC.
References
Wang H, Li Y, Wang Y, Han ZG, Cai B. C9orf100, a new member of the Dbl-family guanine nucleotide exchange factors, promotes cell proliferation and migration in hepatocellular carcinoma. Mol Med Rep. 2012;5:1169–74.
Goicoechea SM, Awadia S, Garcia-Mata R. I’m coming to GEF you: Regulation of RhoGEFs during cell migration. Cell Adh Migr. 2014;8:535–49.
Hennig A, Markwart R, Esparza-Franco MA, Ladds G, Rubio I. Ras activation revisited: role of GEF and GAP systems. Biol Chem. 2015;396:831–48.
Miller NL, Kleinschmidt EG, Schlaepfer DD. RhoGEFs in cell motility: novel links between Rgnef and focal adhesion kinase. Curr Mol Med. 2014;14:221–34.
Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene. 2014;33:4021–35.
Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–80.
Travis WD, Brambilla E, Burke A, Marx A, Nicholson AG. WHO classification of tumours of the lung, pleura, thymus and heart. Lyon: International Agency for Research on Cancer, 2015.
Edge SB, Compton CC. The American Joint Committee on Cancer: the7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–4.
Zhang X, Yu X, Jiang G, et al. Cytosolic TMEM88 promotes invasion and metastasis in lung cancer cells by binding DVLS. Cancer Res. 2015;75:4527–37.
Kruger NJ. The Bradford method for protein quantitation. Basic Protein Pept Protoc. 1994;32:9–15.
Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241.
Wettschureck N, Offermanns S. Rho/Rho-kinase mediated signaling in physiology and pathophysiology. J Mol Med. 2002;80:629–38.
Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–35.
Lopez-Ilasaca M. Signaling from G-protein-coupled receptors to mitogen-activated protein (MAP)-kinase cascades. Biochem Pharmacol. 1998;56:269–77.
Tatsumi E, Yamanaka H, Kobayashi K, Yagi H, Sakagami M, Noguchi K. RhoA/ROCK pathway mediates p38 MAPK activation and morphological changes downstream of P2Y12/13 receptors in spinal microglia in neuropathic pain. Glia. 2015;63:216–28.
Rual JF, Venkatesan K, Hao T, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–8.
Zhu Y, Tian Y, Du J, et al. Dvl2-dependent activation of Daam1 and RhoA regulates Wnt5a-induced breast cancer cell migration. PLoS One. 2012;7:e37823.
Zhu Y, Shen T, Liu J, et al. Rab35 is required for Wnt5a/Dvl2-induced Rac1 activation and cell migration in MCF-7 breast cancer cells. Cell Signal. 2013;25:1075–85.
Acknowledgements
We thank Dr. Hiroshi Kijima and Dr. Gary Bokoch for kind gifts of the LK2 cell line and plasmids. This work was supported by the National Natural Science Foundation of China [grant numbers 81472805 to YM, 81602012 to XZ, 81402520 to AL], the Natural Science Foundation of Liaoning Province, China [grant number 201421044 to YM] and the Research Foundation for the Doctoral Program [grant number 20141040 to AL].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhou, H., Cai, L., Zhang, X. et al. ARHGEF39 promotes tumor progression via activation of Rac1/P38 MAPK/ATF2 signaling and predicts poor prognosis in non-small cell lung cancer patients. Lab Invest 98, 670–681 (2018). https://doi.org/10.1038/s41374-018-0022-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-018-0022-y