Abstract
Mitochondrial nucleoside diphosphate kinase (NDPK-D; synonyms: NME4, NM23-H4) represents the major mitochondrial NDP kinase. The homohexameric complex emerged as a protein with multiple functions in bioenergetics and phospholipid signaling. It occurs at different but precise mitochondrial locations and can affect among other mitochondrial shapes and dynamics, as well as the specific elimination of defective mitochondria or cells via mitophagy or apoptosis. With these various functions in cell homeostasis, NDPK-D/NME4 adds to the group of so-called moonlighting (or gene sharing) proteins.
Similar content being viewed by others
Introduction
Mitochondria generate ATP, and control life and death of the eukaryotic cell. Enzymes in nucleotide conversion are central to relay ATP generated by oxidative phosphorylation to the molecular machineries responsible for mitochondrial functions and dynamics as well as in fine for cellular physiology.
A mitochondrial nucleoside diphosphate kinase (NDPK) activity was described already in 1955 [1], soon after the discovery of the enzyme. NDPKs catalyze the reversible phosphorylation of nucleoside diphosphates into nucleoside triphosphates via a phosphohistidine intermediate and are therefore responsible for cellular NDP/NTP homeostasis [2]. This activity was reported in both the matrix and the extra-matrix compartments, and also associated with the contact sites connecting the inner and outer membranes [3, 4]. The submitochondrial localization was greatly variable, depending on the species and the tissue examined. Mitochondrial NDPK activity was mostly located outside the matrix in pigeon heart, as well as mammalian liver [5]. In addition, NDPK was localized in the matrix in a tissue-dependent manner, in particular in pigeon liver, but also in mammalian heart [5]. In plants [6, 7] and amoeba [8], a mitochondrial NDPK was localized in the intermembrane/cristae space. The membrane-bound or soluble state of the enzyme remained often undefined.
Many functions have been proposed for mitochondrial NDPK. In the matrix, the enzyme was suggested to be associated to the Krebs cycle to synthesize ATP at the expense of GTP provided through succinylthiokinase or, vice versa, to furnish GTP for short chain fatty acid catabolism [9]. In this compartment, it was also proposed to synthesize NTP for protein and nucleic acid synthesis and to play a role in iron homeostasis [10]. In the intermembrane/cristae space, NDPK was proposed to supply NTP at the expense of ATP, synthesized by oxidative phosphorylation, and of NDP, freely diffusible from cytoplasm through the outer membrane [11].
In humans, the only NDPK possessing a typical N-terminal mitochondrial targeting sequence is NDPK-D/NME4 encoded by the NME4 gene located on the short arm of chromosome 16 at 16p13.3, in the same region as NME3 [12, 13]. Two other human genes, NME3 (encoding NDPK-C) and NME6, encode proteins that have been reported to be, at least partly, located to mitochondria [14,15,16]. However, no specific mitochondrial targeting sequence could be detected in these proteins, although NDPK-C/NME3 possesses a N-terminal hydrophobic peptide (17 amino acids), which could anchor the protein to membranes [17, 18]. Finally, a NME1–NME2 read-through fusion protein is annotated as mitochondrial in the MitoProteome database (http://www.mitoproteome.org), but not in MitoCarta2.0. The unique article referring to this protein [19] localizes it to the cytosol, and we neither observed a protein of the corresponding size in mitochondrial extracts of HeLa and HEK cells using anti NDPK-A/NME1 and/or NDPK-B/NME2 antibodies (unpublished data).
NDPK-D/NME4 evolution, expression, and localization
NME4 belongs to the group I members of the NDPK/NME family which can be divided in two groups in mammals, based on phylogenetic analyses [20, 21]. Group I contains NME1, NME2, NME3, and NME4, which share from 58 to 88% identity and are all catalytically active, while group II contains more divergent proteins displaying a single or several NDPK domains associated or not with extra domains (NME5 to 9). A single group I gene ancestor and group II genes (NME5 to 8) originate from duplication events that occurred before the chordate radiation [21]. Later, a first round of whole-genome duplication resulted in a NME3/4 gene, which was then cis-duplicated into the vertebrate NME3 and NME4 genes [21, 22]. NME4 protein has been characterized in human [12], mouse [23], pigeon [24], and Xenopus laevis [25], with a size of 187 amino acids in human [12]. A second mRNA encoding a shorter variant missing the first 70 residues is mentioned in the UniProt data base (NDKM_HUMAN entry). NME4 is addressed to mitochondria through a mitochondria-specific targeting signal, which has to be cleaved to reveal catalytic activity [13] and is not found in any other NME family member [21]. Other known mitochondrial NDPK proteins that possess a canonical mitochondrial targeting sequence are found in amoeba [8], plants [26], and green alga [27]. This feature is thus a likely functional convergence between amoebozoan NDPK, plant NDPKs and vertebrate NME4 [22]. Interestingly, the unique NDPK in yeast lacks a targeting signal and shows dual localization to cytosol and mitochondria [28, 29].
The NME4 gene has been analyzed in some detail. Massé et al. [23] have determined that the mouse and human NME4 genes contain 5 exons, like the other group I enzymes, and span about 3.5 kb. A viable NME4 KO strain is mentioned in the database of the International Mouse Phenotyping Consortium (http://www.mousephenotype.org), but no detailed characterization has been published so far. Human NDPK-D/NME4 is ubiquitously expressed, with mRNA found at high levels in liver, kidney, bladder, prostate, at intermediate levels in heart, colon, intestine, skeletal muscle, and ovary and at low levels in brain, testis, and peripheral leukocytes [12, 30]. In mouse, NME4 mRNA was detected in heart, kidney, and liver in much lower level than the NME1 and NME2 counterparts [31]. In contrast, in adult zebra fish, NME4 mRNA is mainly expressed in gonads, weakly expressed in gills and barely detectable in other tissues [21].
Few studies reported NME4 expression during development and differentiation. NME4 mRNA is expressed at low levels in a few embryonic organs (some brain regions, heart, and intestine) while the other group I members are more widely expressed [31, 32]. From its mRNA distribution, NME4 might have functions distinct from NME1, −2, and −3 and could be mainly involved in cell proliferation during ontogenesis. Tanimura et al. [33] reported that NME4 expression was decreased during differentiation of HL-60 cells to macrophages and neutrophils.
In different human cell lines, endogenous and overexpressed NDPK-D/NME4 wild-type and mutant proteins are all localizing to mitochondria as found by imaging techniques and classical cell fractionation [13, 34, 35]. A nuclear localization of NME4 was recently reported and related to NME4 deacetylation via SIRT1 [32]. Although this secondary modification of NME4 is of interest and the only one published so far, the nuclear localization should be treated with caution. The NDPK-D/NME4 fusion constructs used in these experiments with mouse neuroblastoma cells carry an N-terminal acidic myc-tag just preceding the basic mitochondrial targeting sequence, which could disturb the mitochondrial import mechanism. In our hands, no nuclear NME4 has been observed in various human cell lines, even when overexpressed.
NME4/NDPK D structure, biochemical, and functional characteristics
Eukaryote NDPKs form hexamers, including vertebrate group I enzymes like NDPK-D/NME4 [13] (Fig. 1), while most of the prokaryote enzymes are tetrameric [36, 37]. An interesting structural feature of NDPK-D/NME4 is the serine residue at position 129, which is a proline in all other group I NMEs. This serine is equivalent to the lethal Killer of prune (K-pn) mutation of Drosophila NME (awd) [38] which greatly weakens subunit interactions and hexamer stability [39]. Indeed, NDPK-D/NME4 is highly susceptible to denaturation by heat and urea, and the reverse S129P mutation greatly stabilizes the protein [13]. S129 evolved only more recently, since teleost NDPK-D/NME4s still possess a proline at this position, the serine appearing only in tetrapods after the sarcopterygian radiation [21].
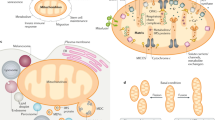
The hexameric structure (PDB 1EHW, monomer backbones in different colors, transparent van der Waals surface in gray) indicating one representative NDP kinase active site (red, phosphotransfer function through phosphorylation of histidine 151) and one basic amino acid triad capable of binding anionic phospholipids, including cardiolipin (blue, phospholipid transfer function). These sites define the two molecular functions of NME4: (i) NTP and ADP generation from mitochondrial ATP and NDPs, and (ii) intermembrane transfer of cardiolipin and other anionic phospholipids by simultaneously binding inner and outer mitochondrial membrane. Via the former function, NME4 in the intermembrane space provides GTP to dynamin-related GTPase OPA1 for mitochondrial dynamics and structure, and ADP to sustain oxidative phosphorylation and mitochondrial respiration, while NME4 in the matrix mainly provides building blocks for mtDNA and RNA synthesis, using ATP but also GTP generated in the Krebs cycle. Via its phospholipid transfer function, NME4 in the intermembrane space facilitates CL transfer to the mitochondrial surface as a pro-mitophagic or pro-apoptotic trigger, and may participate in phospholipid biosynthetic pathways between ER and mitochondria. (Figure modified from Schlattner et al. [72])
NDPK-D/NME4 is bound to the inner mitochondrial membrane mainly through electrostatic interactions [13] between a basic motif (R89-R90-K91) in each monomer and acidic phospholipids, in particular cardiolipin (CL) [34] and possibly some other types of interaction [40]. The R89-R90-K91 motif is located in a surface-exposed loop connecting alpha helices (Fig. 1) and is unique to the NDPK-D/NME4 isoform. CL is synthesized and mainly located at the mitochondrial inner membrane. With its diphosphatidylglycerol structure exposing two negative charges, it could interact with pairs of adjacent basic residues of NDPK-D/NME4 (e.g., R89-R90) that are at a similar distance of about 5–10 Å. Such motifs can be also found in other CL-interacting proteins such as cytochrome c [41] and mitochondrial creatine kinase (mtCK) [42]. This could explain specificity for CL as compared to most other anionic phospholipids [34] and the almost complete loss of membrane interaction when mutating the central arginine 90 (R90D), observed with model liposomes in vitro and mitochondrial membranes in human cervical adenocarcinoma cells (HeLa) in vivo [34]. NDPK-D/NME4 interaction with model membranes also leads to structural alterations of both, the lipid bilayer and the NDPK-D/NME4 hexamer [40]. A basic residue equivalent to R90 can be found in NDPK-D/NME4 ortholog sequences from mammals, fish, and Xenopus, as well as, curiously, in mitochondrial NDPK from Dictyostelium discoideum but not in the mitochondrial NDPK of birds and of lower organisms such as Drosophila, Caenorhabditis elegans, and bacteria [34]. Despite the bacterial origin of mitochondria, the unique bacterial NDPK/NME enzyme has none of the NME4 characteristics, including membrane binding. Mitochondrial NDPK from plants even has an acidic residue at the equivalent site. A R90 equivalent is also absent in the sequences of other human NME protein sequences, and replaced by acidic residues in NDPK-A, -B, and -C (NME1, 2, and 3). The latter creates another type of charged motif that could interact with yet unknown intracellular targets. These data suggest that a specific mitochondrial membrane association appeared late in evolution and could be important for some mitochondrial functions that evolved with vertebrates, including cristae remodeling [43] or certain features of mitophagy, for which NDPK-D/NME4 provides functional support (see below).
NDPK-D/NME4 bound at the inner mitochondrial membrane is preferentially orientated towards the intermembrane space, although some may also face the matrix or be hidden within complexes or cristae structures, as based on the accessibility of enzyme activity before and after detergent lysis of mitochondria [34]. Some further findings support a dual mitochondrial localization of NDPK-D/NME4 in the matrix: it is found associated with the matrix enzyme succinyl-CoA synthetase [44] and mitoribosomes [45], and physically interacts with a transaminase (ABAT) of the mitochondrial nucleoside salvage pathway, leading to mtDNA depletion syndrome when mutated [46]. It is thus conceivable that NDPK-D/NME4 supplies NTP and dNTP for mitochondrial replication and transcription. Finally, Xenopus NDPK-D/NME4 was reported to bind a cytosolic cyclin-dependent kinase inhibitor (p27Xic1) for negative regulation of gliogenesis in the retina. Since these functions would imply the presence of NDPK-D/NME4 in the cytosol [25], further studies on protein localization in Xenopus are required.
Respiration of isolated mitochondria was significantly stimulated by the NDPK substrate TDP in mitochondria containing wild type NDPK-D/NME4, but not in those expressing R90D mutant, which is catalytically equally active. This indicates that optimal functioning of the kinase requires binding to the inner membrane for channeling ADP to ANT (ADP/ATP translocator) and oxidative phosphorylation and vice versa mitochondrial ATP to the NDPK-D/NME4 hexamer [34]. NDPK-D/NME4 not only binds to CL, but also interacts with OPA1, a dynamin-related GTPase bound to the inner membrane and oriented towards the intermembrane space [35]. Interestingly, the wild type enzyme and the kinase-inactive H151N mutant interacted with OPA1 but not the CL-binding-deficient mutant [47]. This interaction provides another type of channeling, namely of GTP and GDP between NDPK-D/NME4 and OPA1 [16, 48], supporting OPA1 functions in cristae shape and mitochondrial fusion and dynamics [49]. These findings emphasize the importance of proper localization NDPK-D/NME4 for its NDP kinase function.
NDPK-D/NME4 and intermembrane phospholipid/cardiolipin transfer
Owing to its symmetrical hexameric structure exposing three R90 on each of its two opposite faces, NDPK-D/NME4 is able to cross-link two membranes containing anionic phospholipids in vitro [50], suggesting that it promotes contact sites between inner and outer mitochondrial membranes in vivo. In this topology, the hexamer is also able to transfer phospholipids between the two membranes, as first shown with liposome model systems in vitro [47]. This requires the presence of CL in the membranes, and does not involve vesicle fusion or loss of the internal contents of the liposomes [50]. Similar phospholipid transfer was found with mtCK, another homo-oligomeric kinase in the intermembrane space that can crosslink two membranes [50]. Interestingly, CL-binding of NDPK-D/NME4 also modulates its NDP kinase activity. Saturation of all CL-binding sites with CL-containing liposomes totally inhibits NDP kinase activity [35], while partial saturation (i.e., the hexamer bound at only at one side to a membrane) only gradually reduces NDP kinase activity (unpublished). This phenomenon is most probably due to the close proximity between CL-binding and catalytic sites (Fig. 1). Some CL-binding of NDPK-D/NME4 seems to be also required for its interaction with OPA1 [47]. The latter, in contrast to NDPK-D/NME4, is strongly stimulated by CL in its GTPase activity [51].
Importantly, NDPK-D/NME4 also facilitates intermembrane transfer of CL in mitochondria in living cells, as first shown in Hela cells by mass spectrometry [35]. CL is synthesized in the inner mitochondrial membrane, leading to high CL concentrations in this membrane, while the CL content of the outer membrane is normally very low [52, 53]. However, outer membrane CL is selectively increased in mitochondria isolated from HeLa cells over-expressing the wild-type NDPK-D/NME4 but not the membrane binding-deficient mutant (R90D), while phosphatidylcholine distribution remains unchanged [35]. Such specific appearance of CL at the mitochondrial surface has been suggested as a signal for triggering apoptosis [54] or mitophagy [55], but underlying molecular mechanisms remained elusive. More recent data now involve NDPK-D/NME4-assisted intermembrane CL transfer in these signaling pathways. HeLa cells expressing the wild-type NDPK-D/NME4, in contrast to those expressing the R90D mutant, were sensitized to rotenone-induced apoptosis [35]. Here, exposure of mostly oxidized CL at the outer membrane [56] leads to recruitment and functional alteration of pro-apoptotic proteins [54, 57]. A similar CL externalization occurs under condition that favor mitophagy, the selective autophagic elimination of damaged mitochondria [55]. The externalized CL, here mostly non-oxidized, is recognized by the microtubule-associated protein 1 light chain 3 (LC3) situated on the surface of the autophagosome. Kagan et al. [47] recently demonstrated that NDPK-D/NME4 is required for mitophagy induced by different mitophagic stimuli in murine lung epithelial MLE-12, SH-SY5Y, and HeLa cells. Overall, NDPK-D/NME4 seems to play a key role in CL signaling for cellular and mitochondrial quality control [47, 58] (see also Schlattner et al. in this issue).
NDPK-D/NME4 in cancer
Most if not all studies on the role of NDPK-D/NME4 in pathology concern cancer, probably because the cytosolic homolog NDPK-A/NME1 is a well studied metastasis suppressor [59]. Still, in contrast to the abundant literature on NME1, only few data are available on NME4 expression in human cancers. Most of them show overexpression of NME4 mRNA in several types of tumors as compared to non-tumor tissue [60, 61] and, in case of gastric cancer, this was predictive for higher patient survival [61]. Other studies report a lower NME4 expression correlating with aggressiveness in a way similar to NME1. In hepatocarcinoma-derived cell lines, NME4 expression was lower in cell lines with high metastatic potential [61]. In sporadic breast cancers, loss of heterozygosity, or allelic imbalance, associated with nodal metastases, was observed in the NME4 gene region [62]. Further, a deletion in the 16p13.3 chromosome region containing the NME4 gene was observed in testicular germ cell tumors of adolescent and young adult men by comparative genomic hybridization microarray with no mention of the mono-allelic or bi-allelic nature of the deletion [63]. In oral cancer, miR-196 was highly overexpressed in cancer tissue and correlated with lymph node metastasis. Functionally, this onco-miR promoted cell migration, invasion and lymph node metastasis through inhibition of NME4 expression, without affecting cell growth [64]. NDPK-D/NME4 was shown to inhibit anchorage-independent growth but not motility of the highly metastatic MDA-MB435 and MDA-MB231 human cell lines [65].
Cancer cells require altered mitochondrial functions, including organelle dynamics, to resist apoptosis and bioenergetic/biosynthetic reprogramming for supporting proliferation, migration, and invasion [66,67,68,69,70]. Since NDPK-D/NME4 plays a role in all of these, it is tempting to speculate that altered expression or function can affect the fate of a cancer cell. Certainly, more targeted research is required to address putative causal links between NDPK-D/NME4 and cell migration, invasion, and metastatic dissemination.
Conclusions
As summarized in this review, mitochondrial NDPK-D/NME4 has different key functions in bioenergetics and lipid signaling and can thus be classified as a moonlighting protein (Fig. 1). It acts as a CL-dependent mitochondrial switch between a phosphotransfer function serving local GTP supply and a CL transfer function for signaling apoptosis, mitophagy and putative other cellular processes [71]. It can fuel GTP to the dynamin-like GTPase OPA1 and thus regulate mitochondrial dynamics. This property is shared by the cytosolic NME1/2 acting on dynamin 1/2 in endocytosis [16]. Further, it supports the concept of NME proteins being part of complexes responsible for channeling NTP for specific cellular processes [48]. Independently of its kinase activity, NDPK-D/NME4 can promote the intermembrane CL transfer to the mitochondrial surface. Under these conditions, it promotes apoptosis to eliminate damaged cells or mitophagy to eliminate damaged mitochondria, both important quality control mechanisms.
Impaired mitochondrial functions participate in the etiology of numerous diseases such as cancer, neurodegeneration and cardiopathy. The available data identify NDPK-D as a key player in the control of cell homeostasis and an important contributor to the quality control of mitochondria. Strategies that aim at favoring or protecting NDPK-D functions may have preventive or therapeutic potential for numerous human pathologies. It is hoped that the availability of invalidated animal models will help to decipher the involved molecular mechanisms.
References
Herbert E, Potter VR, Takagi Y. Nucleotide metabolism. IV. The phosphorylation of 5’-uridine nucleotides by cell fractions from rat liver. J Biol Chem. 1955;213:923–40.
Agarwal RP, Robison B, Parks RE Jr. Nucleoside diphosphokinase from human erythrocytes. Methods Enzymol. 1978;51:376–86.
Adams V, Bosch W, Schlegel J, Wallimann T, Brdiczka D. Further characterization of contact sites from mitochondria of different tissues: topology of peripheral kinases. Biochim Biophys Acta. 1989;981:213–25.
Cherradi N, Defaye G, Chambaz EM. Characterization of the 3 beta-hydroxysteroid dehydrogenase activity associated with bovine adrenocortical mitochondria. Endocrinology. 1994;134:1358–64.
Muhonen WW, Lambeth DO. The compartmentation of nucleoside diphosphate kinase in mitochondria. Comp Biochem Physiol B Biochem Mol Biol. 1995;110:211–23.
Sweetlove LJ, Mowday B, Hebestreit HF, Leaver CJ, Millar AH. Nucleoside diphosphate kinase III is localized to the inter-membrane space in plant mitochondria. FEBS Lett. 2001;508:272–6.
Knorpp C, Johansson M, Baird AM. Plant mitochondrial nucleoside diphosphate kinase is attached to the membrane through interaction with the adenine nucleotide translocator. FEBS Lett. 2003;555:363–6.
Troll H, Winckler T, Lascu I, et al. Separate nuclear genes encode cytosolic and mitochondrial nucleoside diphosphate kinase in Dictyostelium discoideum. J Biol Chem. 1993;268:25469–75.
Krebs HA, Wiggins D. Phosphorylation of adenosine monophosphate in the mitochondrial matrix. Biochem J. 1978;174:297–301.
Gordon DM, Lyver ER, Lesuisse E, Dancis A, Pain D. GTP in the mitochondrial matrix plays a crucial role in organellar iron homoeostasis. Biochem J. 2006;400:163–8.
Pedersen PL. Coupling of adenosine triphosphate formation in mitochondria to the formation of nucleoside triphosphates. Involvement of nucleoside diphosphokinase. J Biol Chem. 1973;248:3956–62.
Milon L, Rousseau-Merck MF, Munier A, et al. nm23-H4, a new member of the family of human nm23/nucleoside diphosphate kinase genes localised on chromosome 16p13. Hum Genet. 1997;99:550–7.
Milon L, Meyer P, Chiadmi M, et al. The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase. J Biol Chem 2000;275:14264–72.
Negroni A, Venturelli D, Tanno B, et al. Neuroblastoma specific effects of DR-nm23 and its mutant forms on differentiation and apoptosis. Cell Death Differ. 2000;7:843–50.
Tsuiki H, Nitta M, Furuya A, et al. A novel human nucleoside diphosphate (NDP) kinase, Nm23-H6, localizes in mitochondria and affects cytokinesis. J Cell Biochem. 1999;76:254–69.
Boissan M, Montagnac G, Shen Q, et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science. 2014;344:1510–5.
Barraud P, Amrein L, Dobremez E, et al. Differential expression of nm23 genes in adult mouse dorsal root ganglia. J Comp Neurol. 2002;444:306–23.
Abu-Taha IH, Heijman J, Hippe HJ, et al. Nucleoside diphosphate kinase-C suppresses cAMP formation in human heart failure. Circulation. 2017;135:881–97.
Valentijn LJ, Koster J, Versteeg R. Read-through transcript from NM23-H1 into the neighboring NM23-H2 gene encodes a novel protein, NM23-LV. Genomics. 2006;87:483–9.
Lacombe ML, Milon L, Munier A, Mehus JG, Lambeth DO. The human Nm23/nucleoside diphosphate kinases. J Bioenerg Biomembr. 2000;32:247–58.
Desvignes T, Pontarotti P, Fauvel C, Bobe J. Nme protein family evolutionary history, a vertebrate perspective. BMC Evol Biol. 2009;9:256.
Desvignes T, Pontarotti P, Bobe J. Nme gene family evolutionary history reveals pre-metazoan origins and high conservation between humans and the sea anemone, Nematostella vectensis. PLoS ONE. 2010;5:e15506.
Masse K, Dabernat S, Bourbon PM, et al. Characterization of the nm23-M2, nm23-M3 and nm23-M4 mouse genes: comparison with their human orthologs. Gene 2002;296:87–97.
Lambeth DO, Mehus JG, Ivey MA, Milavetz BI. Characterization and cloning of a nucleoside-diphosphate kinase targeted to matrix of mitochondria in pigeon. J Biol Chem. 1997;272:24604–11.
Mochizuki T, Bilitou A, Waters CT, Hussain K, Zollo M, Ohnuma S. Xenopus NM23-X4 regulates retinal gliogenesis through interaction with p27Xic1. Neural Dev. 2009;4:1.
Escobar Galvis ML, Hakansson G, Alexciev K, Knorpp C. Cloning and characterisation of a pea mitochondrial NDPK. Biochimie. 1999;81:1089–96.
Anderca MI, Furuichi T, Pinontoan R, Muto S. Identification of a mitochondrial nucleoside diphosphate kinase from the green alga Dunaliella tertiolecta. Plant Cell Physiol. 2002;43:1276–84.
Fukuchi T, Nikawa J, Kimura N, Watanabe K. Isolation, overexpression and disruption of a Saccharomyces cerevisiae YNK gene encoding nucleoside diphosphate kinase. Gene. 1993;129:141–6.
Amutha B, Pain D. Nucleoside diphosphate kinase of Saccharomyces cerevisiae, Ynk1p: localization to the mitochondrial intermembrane space. Biochem J. 2003;370:805–15.
Lacombe ML, Tokarska-Schlattner M, Epand RF, Boissan M, Epand RM, Schlattner U. Interaction of NDPK-D with cardiolipin-containing membranes: structural basis and implications for mitochondrial physiology. Biochimie. 2009;91:779–83.
Amrein L, Barraud P, Daniel JY, Perel Y, Landry M. Expression patterns of nm23 genes during mouse organogenesis. Cell Tissue Res. 2005;322:365–78.
Fujita Y, Fujiwara K, Zenitani S, Yamashita T. Acetylation of NDPK-D regulates its subcellular localization and cell survival. PLoS ONE. 2015;10:e0139616.
Tanimura A, Horiguchi T, Miyoshi K, Hagita H, Noma T. Differential expression of adenine nucleotide converting enzymes in mitochondrial intermembrane space: a potential role of adenylate kinase isozyme 2 in neutrophil differentiation. PLoS ONE. 2014;9:e89916.
Tokarska-Schlattner M, Boissan M, Munier A, et al. The nucleoside diphosphate kinase D (NM23-H4) binds the inner mitochondrial membrane with high affinity to cardiolipin and couples nucleotide transfer with respiration. J Biol Chem. 2008;283:26198–207.
Schlattner U, Tokarska-Schlattner M, Ramirez S, et al. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. J Biol Chem. 2013;288:111–21.
Lascu L, Giartosio A, Ransac S, Erent M. Quaternary structure of nucleoside diphosphate kinases. J Bioenerg Biomembr 2000;32:227–36.
Moynie L, Giraud MF, Georgescauld F, Lascu I, Dautant A. The structure of the Escherichia coli nucleoside diphosphate kinase reveals a new quaternary architecture for this enzyme family. Proteins. 2007;67:755–65.
Biggs J, Tripoulas N, Hersperger E, Dearolf C, Shearn A. Analysis of the lethal interaction between the prune and Killer of prune mutations of Drosophila. Genes Dev. 1988;2:1333–43.
Lascu I, Chaffotte A, Limbourg-Bouchon B, Veron M. A Pro/Ser substitution in nucleoside diphosphate kinase of Drosophila melanogaster (mutation killer of prune) affects stability but not catalytic efficiency of the enzyme. J Biol Chem. 1992;267:12775–81.
Francois-Moutal L, Maniti O, Marcillat O, Granjon T. New insights into lipid-nucleoside diphosphate kinase-D interaction mechanism: protein structural changes and membrane reorganisation. Biochim Biophys Acta. 2013;1828:906–15.
Rytomaa M, Kinnunen PK. Reversibility of the binding of cytochrome c to liposomes. Implications for lipid-protein interactions. J Biol Chem. 1995;270:3197–202.
Schlattner U, Gehring F, Vernoux N, et al. C-terminal lysines determine phospholipid interaction of sarcomeric mitochondrial creatine kinase. J Biol Chem. 2004;279:24334–42.
Olichon A, Elachouri G, Baricault L, Delettre C, Belenguer P, Lenaers G. OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ. 2007;14:682–92.
Kowluru A, Tannous M, Chen HQ. Localization and characterization of the mitochondrial isoform of the nucleoside diphosphate kinase in the pancreatic beta cell: evidence for its complexation with mitochondrial succinyl-CoA synthetase. Arch Biochem Biophys. 2002;398:160–9.
Hensen F, Cansiz S, Gerhold JM, Spelbrink JN. To be or not to be a nucleoid protein: a comparison of mass-spectrometry based approaches in the identification of potential mtDNA-nucleoid associated proteins. Biochimie. 2014;100:219–26.
Besse A, Wu P, Bruni F, et al. The GABA transaminase, ABAT, is essential for mitochondrial nucleoside metabolism. Cell Metab. 2015;21:417–27.
Kagan VE, Jiang J, Huang Z, et al. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ. 2016;23:1140–51.
Zala D, Schlattner U, Desvignes T, et al. The advantage of channeling nucleotides for very processive functions. F1000Res. 2017;6:724.
MacVicar T, Langer T. OPA1 processing in cell death and disease - the long and short of it. J Cell Sci. 2016;129:2297–306.
Epand RF, Schlattner U, Wallimann T, Lacombe ML, Epand RM. Novel lipid transfer property of two mitochondrial proteins that bridge the inner and outer membranes. Biophys J. 2007;92:126–37.
Ban T, Heymann JA, Song Z, Hinshaw JE, Chan DC. OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum Mol Genet. 2010;19:2113–22.
Daum G, Vance JE. Import of lipids into mitochondria. Prog Lipid Res. 1997;36:103–30.
Schlattner U, Tokarska-Schlattner M, Rousseau D, et al. Mitochondrial cardiolipin/phospholipid trafficking: the role of membrane contact site complexes and lipid transfer proteins. Chem Phys Lipids 2014;179:32–41.
Schug ZT, Gottlieb E. Cardiolipin acts as a mitochondrial signalling platform to launch apoptosis. Biochim Biophys Acta. 2009;1788:2022–31.
Chu CT, Ji J, Dagda RK, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–205.
Garcia Fernandez M, Troiano L, Moretti L, et al. Early changes in intramitochondrial cardiolipin distribution during apoptosis. Cell Growth Differ. 2002;13:449–55.
Kagan VE, Chu CT, Tyurina YY, Cheikhi A, Bayir H. Cardiolipin asymmetry, oxidation and signaling. Chem Phys Lipids. 2014;179:64–9.
Maguire JJ, Tyurina YY, Mohammadyani D, et al. Known unknowns of cardiolipin signaling: The best is yet to come. Biochim Biophys Acta. 2017;1862:8–24.
Steeg PS, Bevilacqua G, Kopper L, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–4.
Seifert M, Welter C, Mehraein Y, Seitz G. Expression of the nm23 homologues nm23-H4, nm23-H6, and nm23-H7 in human gastric and colon cancer. J Pathol. 2005;205:623–32.
Xu ZY, Chen JS, Shu YQ. Gene expression profile towards the prediction of patient survival of gastric cancer. Biomed Pharmacother. 2010;64:133–9.
Patocs A, Zhang L, Xu Y, et al. Breast-cancer stromal cells with TP53 mutations and nodal metastases. N Engl J Med. 2007;357:2543–51.
Skotheim RI, Autio R, Lind GE, et al. Novel genomic aberrations in testicular germ cell tumors by array-CGH, and associated gene expression changes. Cell Oncol. 2006;28:315–26.
Lu YC, Chang JT, Liao CT, et al. OncomiR-196 promotes an invasive phenotype in oral cancer through the NME4-JNK-TIMP1-MMP signaling pathway. Mol Cancer 2014;13:218.
McDermott WG, Boissan M, Lacombe ML, Steeg PS, Horak CE. Nm23-H1 homologs suppress tumor cell motility and anchorage independent growth. Clin Exp Metastas. 2008;25:131–8.
Zhao J, Zhang J, Yu M, et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32:4814–24.
Ferreira-da-Silva A, Valacca C, Rios E, et al. Mitochondrial dynamics protein Drp1 is overexpressed in oncocytic thyroid tumors and regulates cancer cell migration. PLoS ONE. 2015;10:e0122308.
Han XJ, Yang ZJ, Jiang LP, et al. Mitochondrial dynamics regulates hypoxia-induced migration and antineoplastic activity of cisplatin in breast cancer cells. Int J Oncol. 2015;46:691–700.
Rehman J, Zhang HJ, Toth PT, et al. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26:2175–86.
Porporato PE, Payen VL, Perez-Escuredo J, et al. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014;8:754–66.
Kagan VE, Tyurina YY, Tyurin VA, et al. Cardiolipin signaling mechanisms: collapse of asymmetry and oxidation. Antioxid Redox Signal 2015;22:1667–80
Schlattner U, Tokarska-Schlattner M, Epand RM, et al. Mitochondrial NM23-H4/NDPK-D: a bifunctional nanoswitch for bioenergetics and lipid signaling. Naunyn Schmiede Arch Pharmacol. 2015;388:271–8.
Acknowledgements
This work was supported by the Fondation pour la Recherche Médicale (FRM; DPM20121125557), the Groupement des Entreprises Françaises contre le Cancer (GEFLUC), Cancéropôle IdF (n° 2015-1-EMERG-25-INSERM 6-1), and a CMIRA Explo’ra doc fellowship of the Region Rhone Alpes.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Lacombe, ML., Tokarska-Schlattner, M., Boissan, M. et al. The mitochondrial nucleoside diphosphate kinase (NDPK-D/NME4), a moonlighting protein for cell homeostasis. Lab Invest 98, 582–588 (2018). https://doi.org/10.1038/s41374-017-0004-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-017-0004-5
This article is cited by
-
NME4 mediates metabolic reprogramming and promotes nonalcoholic fatty liver disease progression
EMBO Reports (2023)
-
The mitochondrially-localized nucleoside diphosphate kinase D (NME4) is a novel metastasis suppressor
BMC Biology (2021)
-
Widely targeted metabolomic analyses unveil the metabolic variations after stable knock-down of NME4 in esophageal squamous cell carcinoma cells
Molecular and Cellular Biochemistry (2020)