Abstract
Objective:
To examine the risk for clinically significant chromosomal microarray analysis (CMA) findings in fetal right aortic arch (RAA).
Methods:
Data from all CMA analyses performed owing to isolated RAA reported to the Israeli Ministry of Health between January 2013 and September 2016 were evaluated retrospectively. Risk for abnormal CMA findings was compared with two control populations, based on both previously described 9272 pregnancies with normal ultrasound, and on a local cohort of 5541 pregnancies undergoing CMA testing owing to maternal request. In addition, Pubmed database search was conducted for original researches examining this issue.
Results:
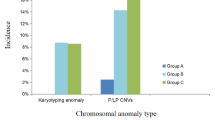
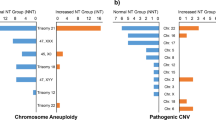
Of 94 CMA analyses performed owing to isolated RAA, six (6.4%) pathogenic findings were detected (47,XX + 21; 45,X; two 22q11.2 microdeletions; 10p15.3 microdeletion and 16p11.2 duplication). Compared with control groups, an isolated RAA yielded a significantly increased relative risk for abnormal CMA results. Literature search yielded two additional retrospective studies describing microarray testing in RAA and encompassing 57 cases. The overall risk for clinically significant CMA findings was 6.62% (10/151).
Conclusions:
CMA testing is indicated in cases of prenatal isolated RAA, even in the era of advanced sonographic equipment, routine biochemical screening for Down syndrome and available non-invasive prenatal testing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Achiron R, Rotstein Z, Heggesh J, Bronshtein M, Zimand S, Lipitz S, et al. Anomalies of the fetal aortic arch: a novel sonographic approach to in-utero diagnosis. Ultrasound Obstet Gynecol. 2002;20:553–557.
Hanneman K, Newman B, Chan F. Congenital variants and anomalies of the aortic arch. Radiographics. 2017;37:32–51.
McElhinney DB, Clark BJ 3rd, Weinberg PM, Kenton ML, McDonald-McGinn D, Driscoll DA, et al. Association of chromosome 22q11 deletion with isolated anomalies of aortic arch laterality and branching. J Am Coll Cardiol. 2001;37:2114–2119.
D’Antonio F, Khalil A, Zidere V, Carvalho JS. Fetuses with right aortic arch: a multicenter cohort study and meta-analysis. Ultrasound Obstet Gynecol. 2016;47:423–432.
American College of Obstetricians and Gynecologists’ Committee on Practice Bulletins—Obstetrics; Committee on Genetics; Society for Maternal–Fetal Medicine. Practice Bulletin No. 162: Prenatal Diagnostic Testing for Genetic Disorders. Obstet Gynecol. 2016;127:e128–e122..
Peng R, Xie HN, Zheng J, Zhou Y, Lin MF. Fetal right aortic arch: associated anomalies, genetic anomalies with chromosomal microarray analysis and postnatal outcome. Prenat Diagn. 2017;37:329–335.
O’Mahony EF, Hutchinson DP, McGillivray G, Nisbet DL, Palma-Dias R. Right-sided aortic arch in the age of microarray. Prenat Diagn. 2017;37:440–445.
Maya I, Kahana S, Yeshaya J, Tenne T, Yacobson S, Agmon-Fishman I, et al. Chromosomal microarray analysis in fetuses with aberrant right subclavian artery. Ultrasound Obstet Gynecol. 2016;49:337–341.
Svirsky R, Reches A, Brabbing-Goldstein D, Bar-Shira A, Yaron Y. Association of aberrant right subclavian artery with abnormal karyotype and microarray results. Prenat Diagn. 2017;37:808–811.
Maya I, Sharony R, Yacobson S, Kahana S, Yeshaya J, Tenne T, et al. When genotype is not predictive of phenotype: implications for genetic counseling based on 21,594 chromosomal microarray analysis examinations. Genet Med. 2017;20:128–131.
Fiorentino F, Caiazzo F, Napolitano S, Spizzichino L, Bono S, Sessa M, et al. Introducing array comparative genomic hybridization into routine prenatal diagnosis practice: a prospective study on over 1000 consecutive clinical cases. Prenat Diagn. 2011;31:1270–1282.
Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST, Working Group of the American College of Medical Genetics Laboratory Quality Assurance C. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–685.
Callaway JL, Shaffer LG, Chitty LS, Rosenfeld JA, Crolla JA. The clinical utility of microarray technologies applied to prenatal cytogenetics in the presence of a normal conventional karyotype: a review of the literature. Prenat Diagn. 2013;33:1119–1123.
Coe BP, Witherspoon K, Rosenfeld JA, van Bon BW, Vulto-van Silfhout AT, Bosco P, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46:1063–1071.
DeScipio C, Conlin L, Rosenfeld J, Tepperberg J, Pasion R, Patel A, et al. Subtelomeric deletion of chromosome 10p15.3: clinical findings and molecular cytogenetic characterization. Am J Med Genet A. 2012;158A:2152–2161.
Vargiami E, Ververi A, Kyriazi M, Papathanasiou E, Gioula G, Gerou S, et al. Severe clinical presentation in monozygotic twins with 10p15.3 microdeletion syndrome. Am J Med Genet A. 2014;164A:764–768.
Eggert M, Muller S, Heinrich U, Mehraein Y. A new familial case of microdeletion syndrome 10p15.3. Eur J Med Genet. 2016;59:179–182.
Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–209.
D’Angelo D, Lebon S, Chen Q, Martin-Brevet S, Snyder LG, Hippolyte L, et al. Defining the effect of the 16p11.2 duplication on cognition, behavior, and medical comorbidities. JAMA Psychiatry. 2016;73:20–30.
Chang H, Li L, Li M, Xiao X. Rare and common variants at 16p11.2 are associated with schizophrenia. Schizophr Res. 2016;184:105–108.
Maillard AM, Ruef A, Pizzagalli F, Migliavacca E, Hippolyte L, Adaszewski S, et al. The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Mol Psychiatry. 2015;20:140–147.
Shinawi M, Liu P, Kang SH, Shen J, Belmont JW, Scott DA, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–341.
Schaaf CP, Goin-Kochel RP, Nowell KP, Hunter JV, Aleck KA, Cox S, et al. Expanding the clinical spectrum of the 16p11.2 chromosomal rearrangements: three patients with syringomyelia. Eur J Human Genet. 2011;19:152–156.
Lazier J, Fruitman D, Lauzon J, Bernier F, Argiropoulos B, Chernos J, et al. Prenatal array comparative genomic hybridization in fetuses with structural cardiac anomalies. J Obstet Gynaecol Can. 2016;38:619–626.
McDonald-McGinn DM, Kirschner R, Goldmuntz E, Sullivan K, Eicher P, Gerdes M, et al. ThePhiladelphia story: the 22q11.2 deletion: report on 250 patients. Genet Couns. 1999;10:11–24.
Bassuk AG, Muthuswamy LB, Boland R, Smith TL, Hulstrand AM, Northrup H, et al. Copy number variation analysis implicates the cell polarity gene glypican 5 as a human spina bifida candidate gene. Hum Mol Genet. 2013;22:1097–111.
Acknowledgements
We are thankful to all the genetic counselors and laboratory personnel who made this data acquisition possible.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Maya, I., Singer, A., Baris, H. et al. Prenatal microarray analysis in right aortic arch—a retrospective cohort study and review of the literature. J Perinatol 38, 468–473 (2018). https://doi.org/10.1038/s41372-018-0062-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41372-018-0062-6