Abstract
Background
Evidence is needed on the presence of SARS-CoV-2 in various types of environmental samples and on the estimated transmission risks in non-healthcare settings on campus.
Objectives
The objective of this research was to collect data on SARS-CoV-2 viral load and to examine potential infection risks of people exposed to the virus in publicly accessible non-healthcare environments on a university campus.
Methods
Air and surface samples were collected using wetted wall cyclone bioaerosol samplers and swab kits, respectively, in a longitudinal environmental surveillance program from August 2020 until April 2021 on the University of Michigan Ann Arbor campus. Quantitative rRT-PCR with primers and probes targeting gene N1 were used for SARS-CoV-2 RNA quantification. The RNA concentrations were used to estimate the probability of infection by quantitative microbial risk assessment modeling and Monte-Carlo simulation.
Results
In total, 256 air samples and 517 surface samples were collected during the study period, among which positive rates were 1.6% and 1.4%, respectively. Point-biserial correlation showed that the total case number on campus was significantly higher in weeks with positive environmental samples than in non-positive weeks (p = 0.001). The estimated probability of infection was about 1 per 100 exposures to SARS-CoV-2-laden aerosols through inhalation and as high as 1 per 100,000 exposures from contacting contaminated surfaces in simulated scenarios.
Significance
Viral shedding was demonstrated by the detection of viral RNA in multiple air and surface samples on a university campus. The low overall positivity rate indicated that the risk of exposure to SARS-CoV-2 at monitored locations was low. Risk modeling results suggest that inhalation is the predominant route of exposure compared to surface contact, which emphasizes the importance of protecting individuals from airborne transmission of SARS-CoV-2 and potentially other respiratory infectious diseases.
Impact
Given the reoccurring epidemics caused by highly infectious respiratory viruses in recent years, our manuscript reinforces the importance of monitoring environmental transmission by the simultaneous sampling and integration of multiple environmental surveillance matrices for modeling and risk assessment.
Similar content being viewed by others
Introduction
On March 11th, 2020, the World Health Organization (WHO) declared Coronavirus Disease 2019 (COVID-19)—a contagious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)—to be a global pandemic [1]. The median incubation time of the virus is 5.1 days post-exposure, with 97.5% of the people who develop symptoms, the most common of which are fever, dry cough, and shortness of breath, doing so within 11.5 days [2]. As of August 2021, the COVID-NET surveillance data from the CDC (Centers for Disease Control and Prevention) shows that, among COVID-19 associated hospitalizations across 14 participating US states, ~22.8% were admitted into intensive care units and roughly 10.2% succumb to their illness [3]. Knowledge of exposure risks is critical to prioritize mitigation strategies to control the spread of this highly infectious respiratory viruses.
The CDC categorizes the transmission modes of SARS-CoV-2 as inhalation of virus, virus deposition on mucous membranes, and contact of mucous membranes with virus-contaminated hands [4]. Based on this information, mask wearing, physical distancing, and contact tracing/quarantine are critical preventive measures for the control of outbreaks until large fractions of the population can be vaccinated against the disease or gain natural immunity through infection.
Studies and experimental results on the environmental persistence of SARS-CoV-2 have indicated the possibility of environmental transmission via surfaces and air in the vicinity of infected persons [5, 6]. Early studies showed that asymptomatic, pre-symptomatic, and symptomatic carriers may shed virus particles, which can remain viable and infectious for hours in aerosol and up to 3 days on surfaces, depending on environmental factors such as temperature, relative humidity, and surface materials [7,8,9]. More recent studies have found that the virus can remain viable on surfaces such as plastic, stainless steel, glass, ceramics, wood, cotton, and paper for 4 to 7 days [7]; however, it may remain viable for longer under appropriate temperatures (<30 °C in experimental conditions) [10]. Understanding the extent of non-clinical environmental transmission, especially through aerosol and fomite (i.e., inanimate objects that are contaminated by infectious agents and can potentially transmit disease to a new host) pathways, has been crucial to tailoring interventions that control the spread of SARS-CoV-2.
University campuses offer a wide range of diverse non-healthcare but potentially risky settings, for example, musical rehearsal studios, performance halls, and indoor intensive physical exercise facilities (i.e., gyms). The exposure risk to SARS-CoV-2 needs to be evaluated in these environments. Aerosol concentrations generated from wind instruments, such as trumpet, oboe, or bass trombones tends to be higher than those generated by normal speaking [11]. In a gym, the sustained heavy breathing during intensive aerobic exercise generates proportionately (to heart rate) higher concentrations of aerosols [12], and mask fit is often intentionally compromised when people have trouble breathing to allow for more air exchange, which can further diminish the effectiveness of face masks in obstructing respiratory jets [13]. Use of shared exercise equipment and public drinking fountains may also spread the virus through surface touch points, given high virus stability on non-porous surface [7]. Given these circumstances, it is important to evaluate the risk of exposure through air and fomites for indoor musical and fitness facilities.
Environmental surveillance represents a straightforward method to assess the exposure to SARS-CoV-2 virus in the air and on surface and can complement other public health surveillance measures (i.e., contact tracing, clinical reports, and laboratory-based testing). However, this strategy is complicated by the limited knowledge in environmental contamination levels for different settings. In this study, we conducted longitudinal air and surface sampling to measure SARS-CoV-2 contamination levels in various campus venues, since aerosols and fomites are considered to be the two major environmental transmission routes for this virus [14]. Our primary goals were to evaluate the presence of viral RNA in aerosols and on surfaces and to describe the relationship between our environmental surveillance results and epidemiological data at the university level. Finally, we estimated the probability of infection inferred from the RNA-positive samples using quantitative microbial risk assessment (QMRA) [15]. Environmental public health surveillance studies such as this have the potential to provide insight on real-life environmental exposure risks to infectious respiratory diseases in preparation for future outbreaks and may represent an additional source of information to support evidence-based decisions on mitigation strategies.
Methods
Sampling sites
Air and surface samples were collected at a variety of non-healthcare settings on the University of Michigan (U-M) Ann Arbor Campus. Locations included classrooms, rehearsal rooms, office areas, cafeterias, buses, gyms, student activity buildings and heating, ventilation and air-conditioning (HVAC) system tunnels. Details on our sampling locations, sampled touch points and the corresponding U-M site specific mitigation strategies are provided in the Supplementary Materials (Sampling Sites Descriptions and Mitigation Strategies sections). Our study began in mid-August 2020, 2 weeks before students returned to campus for the Fall semester and continued through the Fall and Winter semesters (i.e., the end of April 2021).
Environmental-sample collection
Aerosols of 0.5 to 10 μm in diameter were collected using SASS 2300 Wetted Wall Cyclone Samplers (Research International, Inc. Monroe, WA, USA) operating at a flow rate of 325 liters per minute (L/min) [16] and automatically concentrated into 4 milliliters (mL) of distilled water after drawing air for defined sampling times. The collected samples were transferred to a Biosafety Level 2 (BSL-2) microbiology laboratory within 20 min at room temperature for immediate sample processing or stored at −80 °C overnight for next-day analysis. Sampling time varied from 30 min (9750 L of air) to 10 h (195,000 L of air) based on estimated exposure duration, rush hours, and logistical feasibility. Observational notes of indoor temperature and ventilation were recorded when aerosol samples were collected. A summary of sampling time and environmental measurements are provided in the Supplementary Materials (Sampling Site Descriptions). Due to the lack of an appropriately equipped and controlled chamber required for infectious aerosol experiments, we were unable to determine the recovery rate for air samples in our lab. Therefore, we decided to use the recovery rate of 48 ± 10% (mean ± SD) reported in Dybwad et al. [17] where they used a SASS 2300 Wetted Wall Cyclone Sampler to recover bacteriophage MS2 in aerosols.
Surface samples were collected using a wet swabbing technique with 3 M™ Quick Swab 6432 (3M Center, St. Paul, MN, USA) on high-touch areas on the same day as the aerosol samples. Swab samples were kept in the letheen broth provided by the kit and sent to the laboratory following the same handling protocols as air samples’. The recovery rate of the surface swabs used in this study was 51 ± 13% (mean ± SD) evaluated in-house using Human Coronavirus OC43 (HCoV-OC43).
RNA extraction and quantitative rRT-PCR
For each quantification, viral RNA was extracted using a TRIzol reagent method (see Supplementary Materials, RNA Extraction and quantitative rRT-PCR section). Total SARS-CoV-2 viral count was assessed using one-step quantitative real-time reverse transcription-polymerase chain reaction (quantitative rRT-PCR) assay on an Eppendorf Mastercycler® RealPlex2 Real-Time PCR Detection System (Eppendorf, Hamburg, Germany) according to the MIQE guidelines [18]. We used primer/probe sets from the US CDC that target a region of the nucleocapsid (N1) gene (Supplementary Materials, Table S3). Synthetic DNA (VR-3276SD™, ATCC®, Manassas, VA, USA) was used as a PCR positive control and to generate the standard curve. The detailed quantitative rRT-PCR parameters can be found in the Supplementary Materials (Standard Curve and Limit of Detection section). For positive samples, we calculated the genome copy concentration based on the standard curve and converted the detected viral RNA concentration from genome copy per micro liter (gc/μL) of liquid (gc/μL of liquid) into gene copies per liter of air collected (gc/L) or per square centimeter of surface swabbed (gc/cm2).
In addition to calculating viral RNA concentrations for each positive sample, we used reverse Kaplan–Meier method (reverse K-M) to compute the 95th and 98th percentile concentrations in air samples collected from gym rooms in Fall 2020 and Winter 2021, because 90% of the environmental samples from that location were below the limit of detection (i.e., left-censored) and the reverse K-M method does not require assumptions about the distribution of values below the limit of detection [19].
COVID-19 data collection
In order to better understand the association between measured environmental contamination and campus case trend, we linked our data with the verified weekly campus-wide COVID-19 case and testing data from the University of Michigan COVID-19 Dashboard [20]. The weekly case numbers represent positive COVID-19 cases among university students, faculty, staff, and employees identified through the university’s general and occupational health services, athletic department, university COVID-19 sampling and tracking program, and the county health department. COVID-19 cases identified through these methods were verified and cross-checked with the Michigan Disease Surveillance System [21]. Weekly testing data represent the tests—which identified both symptomatic and asymptomatic cases—performed by the various university units listed above.
Statistical analysis
We conducted point-biserial correlation analysis to explore the relationship between campus-wide COVID-19 case number and environmental-sample positivity. Each week during the study period was categorized into two groups: environmental-sample positive or environmental-sample negative. Campus weekly case numbers were paired with environmental samples from all locations, (i.e., gym, office, bus, and classroom). To examine if testing capacity could affect the identification of COVID-19 cases between the two semesters, we implemented Welch two-sample t test and linear regression analysis. All analyses were performed using the open-source statistical package R v.4.1.0. [22].
Quantitative microbial risk assessment (QMRA)
We used QMRA [15] as an analytical approach to estimate the probability of infection after exposure to SARS-CoV-2 when information on the exposure dose and dose–response relationship are available. Briefly, our QMRA involved: (1) creation of two transmission scenarios, i.e., the inhalation of viral-loaded aerosols during exercise [23] and contact with contaminated surfaces [24]; (2) consideration of accounting for transmission loss [25, 26] and infection efficiency [17] to calculate the actual exposure dose, and (3) calculation of the infection probability by incorporating the estimated dose into an exponential dose–response model (parameter k) provided by Watanabe et al. [27]. They used pooled data from transgenic mice susceptible to SARS-CoV and murine hepatitis virus (MHV-1), these data are considered a better estimate of infection probability for SARS-CoV-2 than other microorganisms [28]. The detailed modeling steps and the references for model parameters are summarized in Supplementary Table S4. To merge the aggregated and single parameters into QMRA, we applied Monte-Carlo simulation in R software, with 10,000 replications for each summary parameter. A normal distribution assumption was central to this simulation, although further studies are needed to verify the accuracy of this assumption.
Results
Virus detection in environmental samples
In total, we collected 256 air samples and 517 surface samples from August 2020 to April 2021. Due to temporal changes that could potentially influence campus environmental contamination levels, e.g., campus positive COVID-19 cases, testing capacity, and campus activity intensity, we grouped our study results into two time periods: Fall 2020 (August–December 2020) and Winter 2021 (January–April 2021). A summary of sample results overall and for each semester is shown in Table 1. Overall, 98.4% air samples and 98.5% surface samples were negative for SARS-CoV2 RNA. No positive samples were ever collected from dining halls, rehearsal rooms, performance halls, or student activity buildings.
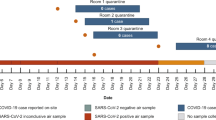
Positive sample details and detected viral RNA concentrations are shown in Table 2. There were four positive air samples and eight positive surface samples in total, presented in Fig. 1 over the on-campus weekly COVID-19 case numbers identified during the sampling period. Sample-positive rates were 1.6% (aerosols) and 1.8% (surface), and 1.4% (aerosols) and 1.1% (surface) for Fall and Winter semesters, respectively. During the Fall 2020 semester, positive air samples were collected from gym rooms (positive/total samples: 2/23) and a campus bus (1/12). Positive surface samples were collected in Fall 2020 from gym drinking fountains and floors (3/76), an office lab (1/12), a classroom student desk surface (1/32), and a campus bus (1/20). During the Winter 2021 semester, viral RNA was detected in each of the following samples: air from a gym weight room (1/48), a gym drinking fountain’s surface (1/143), and surfaces in an office lunchroom (1/6). In gym rooms, the air sample-positive rate dropped from 8.7% to 2.1% between Fall and Winter semesters, and the surface sample-positive rate dropped from 3.9% to 0.7%.
The average temperature measured across locations of positive samples (22 ± 1 °C) was the same as that of measured across locations of negative samples (22 ± 2 °C). There were no significant changes in ventilation systems on the days when positive air samples were detected compared to days when negative samples were collected.
The surface viral RNA concentrations varied from 0.00678 to 0.148 genome copy per square centimeter (gc/cm2). The viral aerosol concentration on the bus was detected at 0.023 genome copy per liter of air (gc/L). Using reverse Kaplan–Meier estimation, the viral aerosol concentration in gym rooms was 0.028 gc/L at the 95th percentile (mean, SE, 95% CI: 0.029, 0.0019, 0.026–0.033) in the fall, and 0.06 gc/L at 98th percentile (mean, SE, 95% CI: 0.029, 0.00098, 0.026–0.033) during the whole study period.
COVID-19 trends and longitudinal environmental surveillance
Point-biserial correlation indicated that in the weeks with positive environmental samples, the campus COVID-19 case numbers were significantly higher than in non-positive weeks (p = 0.001), with means (mean ± standard deviation, SD) of 256 ± 100 and 156 ± 94, respectively (Fig. S2a). No similar trend was found in the building-specific data.
The trends of weekly COVID-19 cases and tests and the relationship between COVID-19 case and test numbers are presented in Fig. S2b. While the Welch two-sample t test showed testing capacity from Fall 2020 to Winter 2021 grew threefold (p < 0.001), weekly campus cases in the two phases were not significantly different (p = 0.98) with an overall mean of 166 ± 103. Linear regression analysis indicated that the weekly COVID-19 case number was significantly positively associated with weekly test number during Fall semester (β = 0.025, p = 0.002), but not the Winter 2021 semester (β = 0.003, p = 0.467).
Probability of infection
We estimated the mean and 95th percentile infection risks for locations with positive viral RNA samples (Table 2). The infection risk through inhalation varied depending on the exposure duration (Fig. 2). For example, after a 40 min moderate- to high- intensity interval training in a gym at the 98th percentile sampled virus concentration level (0.06 gc/L), a mask-less person’s probability of infection would be 15 ± 6 per 1000 exposures through the inhalation of infectious aerosols. On the bus, 15 out of 100,000 mask-less passengers might be infected through inhalation if they take a 5–15 min bus ride when the air on the bus is contaminated by SARS-CoV-2 at the concentration of 0.023 gc/L. Using viral concentration detected on surfaces, the estimated infection risk through fomite transmission was as high as 10−5 (Fig. 2). This level of risk means that after a single touch of the contaminated surface followed by a single finger-to-mucus membrane touch, there is a 1 in 100,000 chance of infection by SARS-CoV-2.
Probability of infection (Pinf, mean with upper bound of 95% confidence interval) for air samples (upper panel) and surface samples (lower panel) derived from Monte-Carlo simulation. Pinf for each sample are presented in chronological order. The tolerable infection risk (dashed lines) is set at 5.5 × 10−4 in Zaneti et al. [28] which can be interpreted as 55 infections per 100,000 exposures.
Discussion
Environmental contamination level in various non-healthcare settings
Several studies have evaluated the existence of SARS-CoV-2 in non-healthcare public places, however, sampling strategies, sample types, and detection primers/probes have varied across studies. Primers/Probes frequently used in rRT-PCR for SARS-CoV-2 virus are E gene [24, 29,30,31,32], N gene [24, 31, 33,34,35,36,37,38], and RdRp/Orf1ab gene [29,30,31,32, 35, 36, 39]. In our study, we decided to use the N target gene as is recommended in the CDC protocol.
As of August 2021, we have found three studies that measured air and/or surface contamination levels in K-12 school settings; viral RNA was detected in multiple samples from classrooms, washrooms, and choir room samples [29, 32, 34]. In our study, a single positive classroom sample was collected from the surface of a student desk, and no air or surface samples from restrooms or choir rooms were positive. In a study by Cordery et al. [32] positive COVID-19 cases were identified after the choir room was found to be contaminated (i.e., positive environmental samples), yet no disease spread was observed following the positive detection. This is consistent with our observation that the environmental transmission efficiency was limited during school musical rehearsal, unlike some documented outbreaks during choir practices where physical distancing was not well implemented during the early stages of the pandemic [40,41,42].
We found that detection of SARS-CoV-2 was most common in gym rooms (75% of the positive air samples and 50% of the positive surface samples). Interestingly, three out of four positive gym surface samples came from the drinking fountain buttons, none came from shared gym equipment. This finding may have resulted from the fact that users of the space were not instructed to clean the water fountain after use as they were with gym equipment (i.e., wipe down sweat as a common courtesy), and so the water fountains were only cleaned occasionally by gym employees. Kozer et al. also detected virus on playground equipment (4.6%) and water fountains (4%) in their study [43]. Nevertheless, the copy concentrations of the positive surface samples were 8 to 1000 times lower than those found in previous studies involving community samples which were positive for E and N1 genes [24, 33], and 10 to 400 times lower than those in healthcare units positive for the N1 gene [33]. The air viral concentration measured in gym rooms was 2500–5000 times lower than that found in hospital air with N1 and N2 primers [37]. Taken together, these finding suggest that viral shedding on this university campus was less than viral shedding in healthcare settings and some public community settings that lack routinely scheduled cleaning and disinfection.
In public service sites, despite substantial anthropic trace contamination on public surfaces [31], surfaces of essential businesses show positive rates ranging from 4.1 to 18.2% using E, N1/N2 or RdRp gene primers [30, 33, 38]. Two studies calculated viral concentration of 1.75 to 16.1 gc/cm2 from public transit surfaces [34], and 0.84 gc/cm2 from bus terminal handrails in comparison with 1.17–39.3 gc/cm2 on healthcare-based surfaces [33]. One study used no-power cold traps to collect air samples into condensed water (1–10 mL) and found viral RNA concentrations that were location dependent, 6.0 gc/mL (3 h concert hall samples), 2.0–5.4 gc/mL (5 h shopping mall samples) [39]. Of the studies that collected air samples in public places, no positive samples were found in outdoor settings [37, 44], while indoor public areas (shopping center, bank, grocery store, markets, and governmental offices, etc.) had a positive rate ranging from 10 to 67%, especially among high traffic flow areas, such as airports, subways, and buses [35, 39]. The higher positivity rates in community settings, compared to the campus results herein, suggest that intervention measures implemented on campus may have helped to reduce environmental transmission.
Throughout the study, no positive air samples were collected from dining halls, rehearsal rooms, performance halls, or student activity buildings. Given the high air flow rate of our sampler and relatively long sampling times, we propose three possible explanations for negative results: (1) the infected persons did not stay long enough to shed a measurable amount of virus; (2) the infected persons were present, but they wore high quality masks properly and disinfected the surfaces after use; (3) aerosols deposited on surfaces or were diluted by the room’s ventilation system before samplers could collect them. Wearing a mask while failing to disinfect the surface may explain why there were days when surface samples were tested positive, yet air samples were negative.
Risk assessment of environmental exposure to SARS-CoV-2
Risk modeling showed higher infection risk through inhalation during intensive exercise without masks than through fomite transmission, which is consistent with CDC’s findings that inhalation is the dominant pathway of spreading the disease as compared to contact with contaminated surfaces [45]. Estimated probability of infection in gym weight rooms (1.45 × 10−2 to 1.84 × 10−2) in this study was similar to modeled probability of infection reported in another fitness center (1.77 × 10−2) [46].
It is equally important to interpret risk with epidemiological data in order to evaluate the public health priorities. In comparing the relative disease burden of SARS-CoV-2, Zaneti et al. developed the tolerable infection risk benchmark of 55 in 100,000 persons based on WHO’s tolerable Disability Adjusted Life Years (DALY) loss per person per year (pppy) and disease/infection ratio observed during the COVID-19 pandemic [28]. The infection risk from exposure to our surface samples in the present study were estimated to be at least 1000 times lower than this tolerable level, while the risk from positive aerosol samples was twenty times higher than the benchmark. The infection risks can be reduced by 87% if a room has 10 air changes per hour (ACH), with recirculation of 36% of air, and filters in its air handling units [46]. The estimated infection risks could be reduced by 38.1 to 98.5% when N95 respirators or available respirator alternatives were used regardless of the ventilation situation [47], and by 49.7% if wearing a cloth mask that has a filtration efficiency of 50% [46]. These findings further reinforce the need to prevent airborne transmission through the implementation of multiple mitigation strategies, such as increased ventilation, air filtration, capacity control, and face coverings, especially considering the risk of breakthrough infections among fully vaccinated people.
A few studies have also adapted QMRA models to quantify infection risk through either aerosol or fomite transmission during the COVID-19 pandemic. Yet, these models have several limitations or need parameter updates. For example, in Harvey et al. [24] the estimated infection risk is between 1 and 40 per 100,000 exposures thru contact with contaminated surfaces. Their exposure dose was calculated using results from positive surface samples collected in an urban community area, where concentrations ranged from 2.54 to 102.43 gc/cm2. At that time, the genome copies to infectious virus ratio (gc:inf, 100 to 1000 gc/PFU) was derived from influenza A (H1N1 and B (H3N2). In our study, we used a gc:inf ratio of 80 gc/PFU based on a SARS-CoV-2 isolation experiment [48], which is more stringent and specific to our QMRA model. Dada and Gyawali [49] and Zaneti et al. [28] used QMRA modeling to estimate the infection risk from occupational exposure of wastewater treatment plant workers through inhalation or accidental ingestion of SARS-CoV-2 virus-contaminated droplets, respectively. Instead of directly measuring the viral aerosol concentration in the air as in our study, they estimated the exposure dose from viral concentration in sewage samples for use in the exponential dose–response equation. Factors such as difference in work type, workers’ proficiency in certain tasks, and the actual aerosolization status could introduce uncertainties for estimating the exposure dose from sewage virus concentration [28]. Moreover, none of the aforementioned studies were able to compare the infection risks between inhalation and surface contact, because: (1) only air or surface samples (but not both) were collected, or (2) when both types of samples were collected in a study, RNA was only detected from either air or surface samples. In this study, aerosols and surface samples were collected simultaneously, analyzed with same methods, and both types of samples had positive detections at the same location (i.e., gym rooms and buses). Therefore, it is possible to compare the risk of infection between the two routes of exposure.
Environmental public health surveillance
As shown in the timeline in Fig. 1, the implementation of several restrictions on social gatherings and other activities were followed by a substantial drop in the weekly case number. In Winter 2021, the weekly case number dropped immediately after COVID-19 testing became mandatory for students at the university following the expansion of testing capacity. A similar drop in the environmental-sample positivity rate was also observed between the two semesters as testing capacity expanded, possibly suggesting an effect of COVID-19 testing on the identification of asymptomatic individuals followed by in-time quarantine and other preventive measures. As testing capacity stabilized, weekly case number reached a low peak in April 2021, which suggested that mandatory testing was an effective but limited mitigation strategy.
Our longitudinal surveillance data shows there were about 100 more COVID-19 weekly cases in environmental-sample-positive weeks than in the weeks with no positive samples, and we suspect that the Fall case number was underreported therefore the case-number gap was likely more than one hundred. Because the positive correlation between weekly case number and testing number became insignificant in the Winter semester may mean that the identification of cases during the Fall semester was limited by testing capacity but this limitation did not continue into the Winter semester due to expanded capacity. Therefore, considering that 75% of the positive samples were detected in Fall, the COVID-19 case number elevation in environmental-sample-positive weeks may have been higher than what was observed in this study, indicating a potential bias toward the null for the findings in this study. Given the correlation between COVID-19 case numbers and the environmental positive samples, our air and surface environmental samples may represent a convenient indicator of virus activity in the community.
Routine, campus-wide environmental sampling could be adopted as a tool for community outbreak surveillance. In Harvey et al. [24] surface samples were shown to serve as a predictive tool, with a 7-day lead time for increases in infections. This suggests the potential use of nowcasting (i.e., estimation of the relatively current situation) or forecasting of outbreaks using comprehensive, routine air and surface surveillance. An approach such as this is a non-invasive way to monitor infectious disease risk in a community, especially where clinical testing resources are scarce. However, we found that COVID-19 cases for specific buildings did not correlate well with our environmental-sample results from those buildings. This is possibly because contact tracing may not identify all positive COVID-19 cases, especially asymptomatic residents, and because random error may cause significant deviation when the positive sample size is small, as is the case when evaluating individual buildings.
In this study, we analyzed SARS-CoV-2 viral RNA in air samples and surface samples collected simultaneously from various public non-healthcare settings on a university campus using quantitative rRT-PCR. The low positivity rate (1.6% and 1.5%, respectively) suggests that the overall risk of environmental transmission in the tested facilities was very limited on campus but more likely in certain locations (i.e., gyms). The estimated risk of infection derived from QMRA modeling revealed higher infection risks from viral aerosol inhalation than from surface contact. This study has several limitations. First, location accessibility, personnel availability, and sample representativeness were heavy considerations during this study and, therefore, limited the scale and frequency of our sampling. Second, given the lockdowns and policies in place to mitigate the spread of COVID-19, no samples were collected among a large aggregation of people, and some samples were collected when only a few people were present, so the negative results need to be interpreted with caution especially as the indoor activities are gradually back to pre-pandemic levels. Third, our study was conducted on a university campus, so extrapolation of these findings to the general population or other non-healthcare settings should be done with caution. Despite these limitations, we believe our results are a valuable addition to the environmental contamination and infectivity data in various non-healthcare settings. The study results support the use of proactive disease mitigation on campuses during this pandemic. The modeling procedures used herein are also valuable in studies of the respiratory infectious diseases with similar transmission mechanisms in preparation for future outbreaks.
Data availability
Observational notes used in this study can be found in Supplementary Files, additional data (e.g., sample notes, lab results, case data) are available from the corresponding author upon reasonable request.
References
Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91:157–60. http://www.ncbi.nlm.nih.gov/pubmed/32191675.
Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. The incubation period of coronavirus disease 2019 (CoVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172:577–82. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7081172/pdf/aim-olf-M200504.pdf.
CDC. Covid-19 Associated hospitalization surveillance network. COVID-NET. 2020. https://gis.cdc.gov/grasp/COVIDNet/COVID19_3.html.
National Center for Immunization and Respiratory Diseases (NCIRD) D of VD. Scientific Brief: SARS-CoV-2 Transmission. CDC COVID-19 Science Briefs. 2021.
Heller L, Mota CR, Greco DB. COVID-19 faecal-oral transmission: are we asking the right questions?. Sci Total Environ. 2020;729:138919. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7182518/pdf/main.pdf.
Pitol AK, Julian TR. Community transmission of SARS-CoV-2 by surfaces: risks and risk reduction strategies. Environ Sci Technol Lett. 2021;8:263–9.
van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–7. https://www.nejm.org/doi/pdf/10.1056/NEJMc2004973?articleTools=true.
Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782–93. https://doi.org/10.1001/jama.2020.12839.
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–62. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7270627/pdf/main.pdf.
Riddell S, Goldie S, Hill A, Eagles D, Drew TW. The effect of temperature on persistence of SARS-CoV-2 on common surfaces. Virol J. 2020;17:1–7.
He R, Gao L, Trifonov M, Hong J. Aerosol generation from different wind instruments. J Aerosol Sci. 2021;151:105669. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7492159/pdf/main.pdf.
Sajgalik P, Garzona-Navas A, Csécs I, Askew JW, Lopez-Jimenez F, Niven AS, et al. Characterization of aerosol generation during various intensities of exercise. Chest. 2021;160:1377–87.
Verma S, Dhanak M, Frankenfield J. Visualizing the effectiveness of face masks in obstructing respiratory jets. Phys Fluids. 2020;32:61708. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7327717/pdf/PHFLE6-000032-061708_1.pdf.
Vardoulakis S, Sheel M, Lal A, Gray D. COVID-19 environmental transmission and preventive public health measures. Aust NZ J Public Health. 2020;44:333–5.
Haas CN. Action levels for SARS-CoV-2 in air: preliminary approach. Risk Anal. 2021;41:705–9. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8251121/pdf/RISA-41-705.pdf.
Guo Z-D, Wang Z-Y, Zhang S-F, Li X, Li L, Li C, et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg Infect Dis. 2020;26:1586.
Dybwad M, Skogan G, Blatny JM. Comparative testing and evaluation of nine different air samplers: end-to-end sampling efficiencies as specific performance measurements for bioaerosol applications. Aerosol Sci Technol. 2014;48:282–95.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22.
Gillespie BW, Chen Q, Reichert H, Franzblau A, Hedgeman E, Lepkowski J, et al. Estimating population distributions when some data are below a limit of detection by using a reverse kaplan-meier estimator. Epidemiology. 2010;21:S64–70.
University of Michigan. U-M COVID-19 Data. Campus Maize & Blueprint. 2020. https://campusblueprint.umich.edu/dashboard/.
Michigan Department of Health & Human Services. Michigan disease surveillance system. 2021. https://www.michigan.gov/mdhhs/0,5885,7-339-71550_5104_31274---,00.html.
R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. https://www.R-project.org.
Gholipour S, Mohammadi F, Nikaeen M, Shamsizadeh Z, Khazeni A, Sahbaei Z, et al. COVID-19 infection risk from exposure to aerosols of wastewater treatment plants. Chemosphere. 2021;273:129701. https://www.ncbi.nlm.nih.gov/pubmed/33517118.
Harvey AP, Fuhrmeister ER, Cantrell ME, Pitol AK, Swarthout JM, Powers JE, et al. Longitudinal monitoring of SARS-CoV-2 RNA on high-touch surfaces in a community setting. Environ Sci Technol Lett. 2021;8:168–75.
Lopez GU, Gerba CP, Tamimi AH, Kitajima M, Maxwell SL, Rose JB. Transfer efficiency of bacteria and viruses from porous and nonporous fomites to fingers under different relative humidity conditions. Appl Environ Microbiol. 2013;79:5728–34. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3754157/pdf/zam5728.pdf.
Pitol AK, Bischel HN, Kohn T, Julian TR. Virus transfer at the skin-liquid interface. Environ Sci Technol. 2017;51:14417–25.
Watanabe T, Bartrand TA, Weir MH, Omura T, Haas CN. Development of a dose-response model for SARS coronavirus. Risk Anal. 2010;30:1129–38. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7169223/pdf/RISA-30-1129.pdf.
Zaneti RN, Girardi V, Spilki FR, Mena K, Westphalen APC, da Costa Colares ER, et al. Quantitative microbial risk assessment of SARS-CoV-2 for workers in wastewater treatment plants. Sci Total Environ. 2021;754:142163.
Crowe J, Schnaubelt AT, Schmidt-Bonne S, Angell K, Bai J, Eske T, et al. Pilot program for test-based SARS-CoV-2 screening and environmental monitoring in an urban public school district. medRxiv. 2021;2021.04.14.21255036. https://www.medrxiv.org/content/10.1101/2021.04.14.21255036v1%0A.
Fernández-de-Mera IG, Rodríguez del-Río FJ, de la Fuente J, Pérez-Sancho M, Hervás D, Moreno I, et al. Detection of environmental SARS-CoV-2 RNA in a high prevalence setting in Spain. Transbound Emerg Dis [Internet]. 2021;68:1487–92. https://www.ncbi.nlm.nih.gov/pubmed/32894654.
Piana A, Colucci ME, Valeriani F, Marcolongo A, Sotgiu G, Pasquarella C, et al. Monitoring COVID-19 Transmission Risks by Quantitative Real-Time PCR Tracing of Droplets in Hospital and Living Environments. mSphere. 2021/01/08. 2021;6. https://www.ncbi.nlm.nih.gov/pubmed/33408231.
Cordery R, Reeves L, Zhou J, Rowan A, Watber P, Rosadas C, et al. Transmission of SARS-CoV-2 by children attending school. Interim report on an observational, longitudinal sampling study of infected children, contacts, and the environment. medRxiv. 2021;2021.03.08.21252839.
Abrahão JS, Sacchetto L, Rezende IM, Rodrigues RAL, Crispim APC, Moura C, et al. Detection of SARS-CoV-2 RNA on public surfaces in a densely populated urban area of Brazil: a potential tool for monitoring the circulation of infected patients. Sci Total Environ. 2020/10/19. 2021;766:142645. https://www.ncbi.nlm.nih.gov/pubmed/33069469.
Roppolo Brazell L, Stetz S, Hipp A, Taylor S, Stark N, Jensen K, et al. Environmental Screening for Surface SARS-CoV-2 Contamination in Urban High-Touch Areas. medRxiv. 2021;2021.05.04.21256107. http://medrxiv.org/content/early/2021/05/04/2021.05.04.21256107.
Hadei M, Mohebbi SR, Hopke PK, Shahsavani A, Bazzazpour S, Alipour M, et al. Presence of SARS-CoV-2 in the air of public places and transportation. Atmos Pollut Res. 2021;12:302–6. https://www.ncbi.nlm.nih.gov/pubmed/33519256.
Kayalar Ö, Arı A, Babuççu G, Konyalılar N, Doğan Ö, Can F, et al. Existence of SARS-CoV-2 RNA on ambient particulate matter samples: a nationwide study in Turkey. Sci Total Environ. 2021;789:147976. https://www.ncbi.nlm.nih.gov/pubmed/34058581.
Passos RG, Silveira MB, Abrahão JS. Exploratory assessment of the occurrence of SARS-CoV-2 in aerosols in hospital facilities and public spaces of a metropolitan center in Brazil. Environ Res. 2021;195:110808. https://www.ncbi.nlm.nih.gov/pubmed/33513382.
Silva SJ, Nascimento J, Reis W, Silva C, Silva P, Mendes R, et al. Widespread Contamination of SARS-CoV-2 on Highly Touched Surfaces in Brazil During the Second Wave of the COVID-19 Pandemic. medRxiv [Internet]. 2021;2021.06.14.21258894. http://medrxiv.org/content/early/2021/06/21/2021.06.14.21258894.abstract.
Gehrke SG, Förderer C, Stremmel W. SARS-CoV-2 airborne surveillance using non-powered cold traps. medRxiv. 2021;1–17. https://www.medrxiv.org/content/medrxiv/early/2021/01/26/2021.01.19.21250064.full.pdf.
Charlotte N. High rate of SARS-CoV-2 transmission due to choir practice in France at the beginning of the COVID-19 pandemic. J Voice. 2020;S0892-1997:30452–5.
Hamner L, Dubbel P, Capron I, Ross A, Jordan A, Lee J, et al. High SARS-CoV-2 attack rate following exposure at a choir practice—Skagit County, Washington, March 2020. MMWR Morbidity Mortal Wkly Rep. 2020;69:606–10.
Richter B, Hipp A, Schubert B, Axt MR, Stratmann M, Schmölder C, et al. From classic to rap: Airborne transmission of different singing styles, with respect to risk assessment of a SARS-CoV-2 infection. medRxiv. 2021. https://www.medrxiv.org/content/early/2021/03/26/2021.03.25.21253694.
Kozer E, Rinott E, Kozer G, Bar-Haim A, Benveniste-Levkovitz P, Klainer H, et al. Presence of SARS-CoV-2 RNA on playground surfaces and water fountains. Epidemiol Infect. 2021;149:e67. https://www.ncbi.nlm.nih.gov/pubmed/33678202.
Chirizzi D, Conte M, Feltracco M, Dinoi A, Gregoris E, Barbaro E, et al. SARS-CoV-2 concentrations and virus-laden aerosol size distributions in outdoor air in north and south of Italy. Environ Int. 2021;146:106255. https://www.ncbi.nlm.nih.gov/pubmed/33221596.
National Center for Immunization and Respiratory Diseases (NCIRD) D of VD. Science Brief: SARS-CoV-2 and Surface (Fomite) Transmission for Indoor Community Environments. CDC COVID-19 Science Briefs. 2021. https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/surface-transmission.html.
Li H, Shankar SN, Witanachchi CT, Lednicky JA, Loeb JC, Alam MM, et al. Environmental surveillance and transmission risk assessments for SARS-CoV-2 in a fitness center. Aerosol Air Qual Res. 2021;21:210106.
Sickbert-Bennett EE, Samet JM, Clapp PW, Chen H, Berntsen J, Zeman KL, et al. Filtration efficiency of hospital face mask alternatives available for use during the COVID-19 pandemic. JAMA Intern Med. 2020;180:1607–12.
Schijven J, Vermeulen LC, Swart A, Meijer A, Duizer E, de Roda Husman AM. Quantitative microbial risk assessment for airborne transmission of sars-cov-2 via breathing, speaking, singing, coughing, and sneezing. Environ Health Perspect. 2021;129:47002.
Dada AC, Gyawali P. Quantitative microbial risk assessment (QMRA) of occupational exposure to SARS-CoV-2 in wastewater treatment plants. Sci Total Enviro. 2021;763:142163. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7560119/pdf/main.pdf.
Acknowledgements
We would like to show our gratitude to the University of Michigan for funding this research. We thank Jon Lillemoen and Philip Szornyi in U-M Environment, Health & Safety Department for their support in coordinating on-campus sampling. We thank Gyan Rusconi-Rodrigues for his assistance in the field sampling, and we also thank Abas Shkembi for his analytical support on the Monte-Carlo simulation.
Funding
Funding for this study was provided by the University of Michigan.
Author information
Authors and Affiliations
Contributions
AF, JD, CX, and RN designed the study. XZ, JW, LS, OY, and XL obtained the data. XZ performed the statistical analysis. XZ and JW drafted the paper. JW, LS, AF, JD, CX, and RN review and edited the paper. All authors read and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, X., Wu, J., Smith, L.M. et al. Monitoring SARS-CoV-2 in air and on surfaces and estimating infection risk in buildings and buses on a university campus. J Expo Sci Environ Epidemiol 32, 751–758 (2022). https://doi.org/10.1038/s41370-022-00442-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41370-022-00442-9
Keywords
This article is cited by
-
Environmental surface surveillance during a COVID-19 outbreak in a school community: implications for controlling indirect transmission
Brazilian Journal of Microbiology (2024)
-
A Wells-Riley based COVID-19 infectious risk assessment model combining both short range and room scale effects
Building Simulation (2024)
-
A coupled experimental and statistical approach for an assessment of SARS-CoV-2 infection risk at indoor event locations
BMC Public Health (2023)
-
Intranasal mask for protecting the respiratory tract against viral aerosols
Nature Communications (2023)
-
Resolving the “health vs environment” dilemma with sustainable disinfection during the COVID-19 pandemic
Environmental Science and Pollution Research (2023)