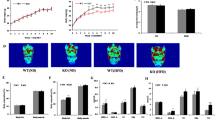
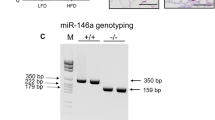
Abstract
Obesity and substantially increased risk of metabolic diseases have become a global epidemic. microRNAs have attracted a great deal of attention as a potential therapeutic target for obesity. MiR-143 has been known to specifically promote adipocyte differentiation by downregulating extracellular signal-regulated kinase 5. Our latest study found that miR-143 knockout is against diet-induced obesity by promoting brown adipose tissue thermogenesis and inhibiting white adipose tissue adipogenesis. Moreover, LPS- or IL-6-induced inhibition of miR-143 expression in brown adipocytes promotes thermogenesis by targeting adenylate cyclase 9. In this review, we will summarize the expression and functions of miR-143 in different tissues, the influence of obesity on miR-143 in various tissues, the important role of adipose-derived miR-143 in the development of obesity, the role of miR-143 in immune cells and thermoregulation and discuss the potential significance and application prospects of miR-143 in obesity management.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bluher M. Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol. 2019;15:288–98.
Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–6.
Yancy WS Jr, Olsen MK, Guyton JR, Bakst RP, Westman EC. A low-carbohydrate, ketogenic diet versus a low-fat diet to treat obesity and hyperlipidemia: a randomized, controlled trial. Ann Intern Med. 2004;140:769–77.
Piche ME, Tchernof A, Despres JP. Obesity phenotypes, diabetes, and cardiovascular diseases. Circ Res. 2020;126:1477–500.
Brown KA. Metabolic pathways in obesity-related breast cancer. Nat Rev Endocrinol. 2021;17:350–63.
Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18:181.
Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–17.
van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–8.
Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–76.
Chen Y, Peng H, Zhou S, Zhuang Y. ADAR1 is targeted by miR-143 to regulate IL-1beta-induced endothelial activation through the NFkappaB pathway. Int J Biochem Cell Biol. 2017;89:25–33.
Pfeifer A, Hoffmann LS. Brown, beige, and white: the new color code of fat and its pharmacological implications. Annu Rev Pharmacol Toxicol. 2015;55:207–27.
Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–5.
Yi C, Xie WD, Li F, Lv Q, He J, Wu J, et al. MiR-143 enhances adipogenic differentiation of 3T3-L1 cells through targeting the coding region of mouse pleiotrophin. FEBS Lett. 2011;585:3303–9.
Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–7.
Bae IS, Park PJ, Lee JH, Cho EG, Lee TR, Kim SH. PPARgamma-mediated G-protein coupled receptor 120 signaling pathway promotes transcriptional activation of miR-143 in adipocytes. Gene. 2017;626:64–9.
Kajimoto K, Naraba H, Iwai N. MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA. 2006;12:1626–32.
Chen L, Hou J, Ye L, Chen Y, Cui J, Tian W, et al. MicroRNA-143 regulates adipogenesis by modulating the MAP2K5-ERK5 signaling. Sci Rep. 2014;4:3819.
Jordan SD, Kruger M, Willmes DM, Redemann N, Wunderlich FT, Bronneke HS, et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011;13:434–46.
Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH, Liang S, et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012;31:1985–98.
Fang R, Xiao T, Fang Z, Sun Y, Li F, Gao Y, et al. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J Biol Chem. 2012;287:23227–35.
Peschiaroli A, Giacobbe A, Formosa A, Markert EK, Bongiorno-Borbone L, Levine AJ, et al. miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene. 2013;32:797–802.
Miao Y, Zhang LF, Guo R, Liang S, Zhang M, Shi S. et al. (18)F-FDG PET/CT for monitoring the response of breast cancer to miR-143-based therapeutics by targeting tumor glycolysis. Mol Ther Nucleic Acids. 2016;5:e357
Chen J, Yu Y, Li H, Hu Q, Chen X, He Y, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. 2019;18:33.
Wang Y, Li G, Zhao L, Lv J. Long noncoding RNA HOTTIP alleviates oxygen-glucose deprivation-induced neuronal injury via modulating miR-143/hexokinase 2 pathway. J Cell Biochem. 2018;119:10107–17.
Zeng XZ, Liu N, Zhang J, Wang L, Zhang ZC, Zhu J, et al. Inhibition of miR-143 during ischemia cerebral injury protects neurones through recovery of the hexokinase 2-mediated glucose uptake. Biosci Rep. 2017;37:BSR20170216.
Muralimanoharan S, Maloyan A, Myatt L. Mitochondrial function and glucose metabolism in the placenta with gestational diabetes mellitus: role of miR-143. Clin Sci (Lond). 2016;130:931–41.
Fong LY, Jing R, Smalley KJ, Taccioli C, Fahrmann J, Barupal DK, et al. Integration of metabolomics, transcriptomics, and microRNA expression profiling reveals a miR-143-HK2-glucose network underlying zinc-deficiency-associated esophageal neoplasia. Oncotarget. 2017;8:81910–25.
Xu RH, Liu B, Wu JD, Yan YY, Wang JN. miR-143 is involved in endothelial cell dysfunction through suppression of glycolysis and correlated with atherosclerotic plaques formation. Eur Rev Med Pharmacol Sci. 2016;20:4063–71.
Miao Y, Zhang LF, Zhang M, Guo R, Liu MF, Li B. Therapeutic delivery of miR-143 targeting tumor metabolism in poorly differentiated thyroid cancer xenografts and efficacy evaluation using F-18-FDG microPET-CT. Hum Gene Ther. 2019;30:882–92.
Yao MY, Wang XH, Tang Y, Zhang WH, Cui B, Liu QH, et al. Dicer mediating the expression of miR-143 and miR-155 regulates hexokinase II associated cellular response to hypoxia. Am J Physiol Lung C. 2014;307:L829–L37.
Sun XH, Zhang L. MicroRNA-143 suppresses oral squamous cell carcinoma cell growth, invasion and glucose metabolism through targeting hexokinase 2. Biosci Rep. 2017;37:BSR20160404.
Guo Y, Liang F, Zhao FL, Zhao J. Resibufogenin suppresses tumor growth and Warburg effect through regulating miR-143-3p/HK2 axis in breast cancer. Mol Cell Biochem. 2020;466:103–15.
Gregersen LH, Jacobsen A, Frankel LB, Wen JY, Krogh A, Lund AH. MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC Cancer. 2012;12:232.
Zhou P, Chen WG, Li XW. MicroRNA-143 acts as a tumor suppressor by targeting hexokinase 2 in human prostate cancer. Am J Cancer Res. 2015;5:2056–63.
Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–10.
Chivukula RR, Shi G, Acharya A, Mills EW, Zeitels LR, Anandam JL, et al. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell. 2014;157:1104–16.
Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903.
Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31.
Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–9.
Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–47.
Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–8.
Kent OA, McCall MN, Cornish TC, Halushka MK. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res. 2014;42:7528–38.
Zhong Y, Li L, Chen Z, Diao S, He Y, Zhang Z, et al. MIR143 inhibits steroidogenesis and induces apoptosis repressed by H3K27me3 in granulosa cells. Front Cell Dev Biol. 2020;8:565261.
Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–78.
Sun K, Kusminski CM, Scherer PE. Adipose tissue remodeling and obesity. J Clin Invest. 2011;121:2094–101.
Vishvanath L, Gupta RK. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. J Clin Invest. 2019;129:4022–31.
Zhang L, Wu ZQ, Wang YJ, Wang M, Yang WC. MiR-143 regulates milk fat synthesis by targeting Smad3 in bovine mammary epithelial cells. Animals (Basel). 2020;10:1453.
Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol R. 2011;75:50–83.
Hu E, Kim JB, Sarraf P, Spiegelman BM. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARγ. Science. 1996;274:100–3.
Akaike M, Che W, Marmarosh NL, Ohta S, Osawa M, Ding B, et al. The hinge-helix 1 region of peroxisome proliferator-activated receptor gamma1 (PPARgamma1) mediates interaction with extracellular signal-regulated kinase 5 and PPARgamma1 transcriptional activation: involvement in flow-induced PPARgamma activation in endothelial cells. Mol Cell Biol. 2004;24:8691–704.
Erding Hu JBK, Sarraf P, Spiegelman BM. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARgamma. Science. 1996;274:2100–3.
Festuccia WT, Laplante M, Berthiaume M, Gelinas Y, Deshaies Y. PPARgamma agonism increases rat adipose tissue lipolysis, expression of glyceride lipases, and the response of lipolysis to hormonal control. Diabetologia. 2006;49:2427–36.
Hua TNM, Kim MK, Vo VTA, Choi JW, Choi JH, Kim HW, et al. Inhibition of oncogenic Src induces FABP4-mediated lipolysis via PPARgamma activation exerting cancer growth suppression. EBioMedicine. 2019;41:134–45.
Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, et al. PPAR gamma signaling and metabolism: the good, the bad and the future. Nat Med. 2013;19:557–66.
Camp HS, Tafuri SR. Regulation of peroxisome proliferator-activated receptor gamma activity by mitogen-activated protein kinase. J Biol Chem. 1997;272:10811–6.
Yang PB, Hou PP, Liu FY, Hong WB, Chen HZ, Sun XY, et al. Blocking PPARgamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression. Proc Natl Acad Sci USA. 2020;117:27412–22.
Liu J, Liu J, Zeng D, Wang H, Wang Y, Xiong J, et al. MiR-143-null is against diet-induced obesity by promoting BAT thermogenesis and inhibiting WAT adipogenesis. Int J Mol Sci. 2022;23:13058.
Li B, Fan J, Chen N. A novel regulator of type II diabetes: microRNA-143. Trends Endocrinol Metab. 2018;29:380–8.
Chen X, Luo J, Yang L, Guo Y, Fan Y, Liu J, et al. miR-143-mediated responses to betaine supplement repress lipogenesis and hepatic gluconeogenesis by targeting MAT1a and MAPK11. J Agric Food Chem. 2022;70:7981–992.
Ji YX, Wang YT, Li PL, Cai L, Wang XM, Bai L, et al. A kinome screen reveals that Nemo-like kinase is a key suppressor of hepatic gluconeogenesis. Cell Metab. 2021;33:1171.
Xu PZ, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–5.
Xihua L, Shengjie T, Weiwei G, Matro E, Tingting T, Lin L, et al. Circulating miR-143-3p inhibition protects against insulin resistance in metabolic syndrome via targeting of the insulin-like growth factor 2 receptor. Transl Res. 2019;205:33–43.
Dahlman I, Belarbi Y, Laurencikiene J, Pettersson AM, Arner P, Kulyte A. Comprehensive functional screening of miRNAs involved in fat cell insulin sensitivity among women. Am J Physiol Endocrinol Metab. 2017;312:E482–94.
Zhang P, Du J, Wang L, Niu L, Zhao Y, Tang G, et al. MicroRNA-143a-3p modulates preadipocyte proliferation and differentiation by targeting MAPK7. Biomed Pharmacother. 2018;108:531–9.
Takanabe R, Ono K, Abe Y, Takaya T, Horie T, Wada H, et al. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem Biophys Res Commun. 2008;376:728–32.
Li M, Wu H, Luo Z, Xia Y, Guan J, Wang T, et al. An atlas of DNA methylomes in porcine adipose and muscle tissues. Nat Commun. 2012;3:850.
Nazari M, Saberi A, Karandish M, Neisi N, Jalali MT, Makvandi M. Influence of L-carnitine on the expression level of adipose tissue miRNAs related to weight changes in obese rats. Pak J Biol Sci. 2016;19:227–32.
Nadler ST, Stoehr JP, Schueler KL, Tanimoto G, Yandell BS, Attie AD. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc Natl Acad Sci USA. 2000;97:11371–6.
van den Berghe G. The role of the liver in metabolic homeostasis: implications for inborn errors of metabolism. J Inherit Metab Dis. 1991;14:407–20.
Fernandes T, Hashimoto NY, Magalhaes FC, Fernandes FB, Casarini DE, Carmona AK, et al. Aerobic exercise training-induced left ventricular hypertrophy involves regulatory MicroRNAs, decreased angiotensin-converting enzyme-angiotensin ii, and synergistic regulation of angiotensin-converting enzyme 2-angiotensin (1-7). Hypertension. 2011;58:182–9.
de Gonzalo-Calvo D, Davalos A, Montero A, Garcia-Gonzalez A, Tyshkovska I, Gonzalez-Medina A, et al. Circulating inflammatory miRNA signature in response to different doses of aerobic exercise. J Appl Physiol (1985). 2015;119:124–34.
Mori MA, Ludwig RG, Garcia-Martin R, Brandao BB, Kahn CR. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 2019;30:656–73.
Ji C, Guo X. The clinical potential of circulating microRNAs in obesity. Nat Rev Endocrinol. 2019;15:731–43.
Al-Rawaf HA. Circulating microRNAs and adipokines as markers of metabolic syndrome in adolescents with obesity. Clin Nutr. 2019;38:2231–8.
Vatandoost N, Amini M, Iraj B, Momenzadeh S, Salehi R. Dysregulated miR-103 and miR-143 expression in peripheral blood mononuclear cells from induced prediabetes and type 2 diabetes rats. Gene. 2015;572:95–100.
Can U, Buyukinan M, Yerlikaya FH. The investigation of circulating microRNAs associated with lipid metabolism in childhood obesity. Pediatr Obes. 2016;11:228–34.
Kilic ID, Dodurga Y, Uludag B, Alihanoglu YI, Yildiz BS, Enli Y, et al. MicroRNA -143 and -223 in obesity. Gene. 2015;560:140–2.
Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:6478.
van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–28.
Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–8.
Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–79.
Chen S, Tang Y, Liu Y, Zhang P, Lv L, Zhang X, et al. Exosomes derived from miR-375-overexpressing human adipose mesenchymal stem cells promote bone regeneration. Cell Prolif. 2019;52:e12669.
Ying W, Riopel M, Bandyopadhyay G, Dong Y, Olefsky JM. Adipose tissue macrophage-derived exosomal miRNAs can modulate invivo and invitro insulin sensitivity. Cell. 2017;171:372.
Isabel H-D, Chen-Yu Z, Antonio V-P. Extracellular vesicles: novel mediators of cell communication in metabolic disease. Trends Endocrinol Metab. 2017;28:3–18.
Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–5.
De Ng ZB, Poliakov A, Hardy RW, Clements R, Zhang HG. Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes. 2009;58:2498–505.
Zhou X, Li Z, Qi M, Zhao P, Duan Y, Yang G, et al. Brown adipose tissue-derived exosomes mitigate the metabolic syndrome in high fat diet mice. Theranostics. 2020;10:8197.
Engin AB. MicroRNA and adipogenesis. Adv Exp Med Biol. 2017;960:489–509.
Lee MS, Kim Y. Mulberry fruit extract ameliorates adipogenesis via increasing AMPK activity and downregulating microRNA-21/143 in 3T3-L1 adipocytes. J Med Food. 2020;23:266–72.
Calo N, Ramadori P, Sobolewski C, Romero Y, Maeder C, Fournier M, et al. Stress-activated miR-21/miR-21* in hepatocytes promotes lipid and glucose metabolic disorders associated with high-fat diet consumption. Gut. 2016;65:1871–81.
Shamsi F, Wang CH, Tseng YH. The evolving view of thermogenic adipocytes – ontogeny, niche and function. Nat Rev Endocrinol. 2021;17:726–44.
Sun W, Modica S, Dong H, Wolfrum C. Plasticity and heterogeneity of thermogenic adipose tissue. Nat Metab. 2021;3:751–61.
Liu J, Zeng D, Luo J, Wang H, Xiong J, Chen X, et al. LPS-induced inhibition of miR-143 expression in brown adipocytes promotes thermogenesis and fever. Int J Mol Sci. 2022;23:13805.
Sunahara RK, Dessauer CW, Gilman AG. Complexity and diversity of mammalian adenylyl cyclases. Annu Rev Pharmacol Toxicol. 1996;36:461–80.
Lazar AM, Irannejad R, Baldwin TA, Sundaram AB, Gutkind JS, Inoue A, et al. G protein-regulated endocytic trafficking of adenylyl cyclase type 9. Elife. 2020;9:e58039–53.
Qi C, Sorrentino S, Medalia O, Korkhov VM. The structure of a membrane adenylyl cyclase bound to an activated stimulatory G protein. Science. 2019;364:389–94.
Blondin DP, Nielsen S, Kuipers EN, Severinsen MC, Jensen VH, Miard S, et al. Human brown adipocyte thermogenesis is driven by beta2-AR stimulation. Cell Metab. 2020;32:287–300.
Rautureau Y, Deschambault V, Higgins ME, Rivas D, Mecteau M, Geoffroy P, et al. ADCY9 (Adenylate Cyclase Type 9) inactivation protects from atherosclerosis only in the absence of CETP (Cholesteryl Ester Transfer Protein). Circulation. 2018;138:1677–92.
Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19:121–35.
Garcia D, Shaw RJ. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol Cell. 2017;66:789–800.
Trefts E, Shaw RJ. AMPK: restoring metabolic homeostasis over space and time. Mol Cell. 2021;81:3677–90.
Scheele C, Wolfrum C. Brown adipose crosstalk in tissue plasticity and human metabolism. Endocr Rev. 2020;41:53–65.
Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18:363–74.
Brestoff JR, Artis D. Immune regulation of metabolic homeostasis in health and disease. Cell. 2015;161:146–60.
Villarroya F, Cereijo R, Villarroya J, Giralt M. Brown adipose tissue as a secretory organ. Nat Rev Endocrinol. 2017;13:26–35.
Yang FT, Stanford KI. Batokines: mediators of inter-tissue communication (a mini-review). Curr Obes Rep. 2022;11:1–9.
Zhang T, Zhang Z, Li F, Ping Y, Qin G, Zhang C, et al. miR-143 regulates memory T cell differentiation by reprogramming T cell metabolism. J Immunol. 2018;201:2165–75.
Qiu X, Shi Q, Huang Y, Jiang H, Qin S. miR-143/145 inhibits Th9 cell differentiation by targeting NFATc1. Mol Immunol. 2021;132:184–91.
Arany Z. Taking a BAT to the chains of diabetes. N Engl J Med. 2019;381:2270–2.
He L, Tang M, Xiao T, Liu H, Liu W, Li G, et al. Obesity-associated miR-199a/214 cluster inhibits adipose browning via PRDM16-PGC-1alpha transcriptional network. Diabetes. 2018;67:2585–600.
Fischer C, Seki T, Lim S, Nakamura M, Andersson P, Yang Y, et al. A miR-327-FGF10-FGFR2-mediated autocrine signaling mechanism controls white fat browning. Nat Commun. 2017;8:2079.
Chen Y, Buyel JJ, Hanssen MJ, Siegel F, Pan R, Naumann J, et al. Exosomal microRNA miR-92a concentration in serum reflects human brown fat activity. Nat Commun. 2016;7:11420.
Wong JJ, Au AY, Gao D, Pinello N, Kwok CT, Thoeng A, et al. RBM3 regulates temperature sensitive miR-142-5p and miR-143 (thermomiRs), which target immune genes and control fever. Nucleic Acids Res. 2016;44:2888–97.
Wang Q, Li D, Cao G, Shi Q, Zhu J, Zhang M, et al. IL-27 signalling promotes adipocyte thermogenesis and energy expenditure. Nature. 2021;600:314–8.
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006.
Heneghan HM, Miller N, McAnena OJ, O’Brien T, Kerin MJ. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J Clin Endocrinol Metab. 2011;96:E846–50.
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54.
US National Library of Medicine. ClinicalTrials.gov. 2016. https://clinicaltrials.gov/ct2/show/NCT02826525.
US National Library of Medicine. ClinicalTrials.gov. 2015. https://clinicaltrials.gov/ct2/show/NCT02612662.
Acknowledgements
The research was supported by grants from the National Natural Science Foundation of China (32072812, 31802156, 32072814), and the Key Project of Guangdong Provincial Nature Science Foundation (2021A1515011310, 2020A1515010062).
Author information
Authors and Affiliations
Contributions
JL, HW, and DZ drafted the manuscript. JX, JL, XC, TC, QX, and JS collected the data and organized the references. XR and YZ participated in the study design. All of the authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, J., Wang, H., Zeng, D. et al. The novel importance of miR-143 in obesity regulation. Int J Obes 47, 100–108 (2023). https://doi.org/10.1038/s41366-022-01245-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41366-022-01245-6