Abstract
Background/objectives
Pregnancies complicated by gestational diabetes (GDM) or maternal obesity have been linked to the development of diabetes, obesity, and fatty liver disease later in life with sex-specific manifestations. Alterations in miRNA expression in offspring exposed to GDM and maternal obesity and effects on hepatic development are unknown. Here, we describe how exposure to maternal obesity in utero leads to sex-specific changes in miRNA and target gene expression in human fetal liver.
Methods
Candidate miRNA expression was measured in second trimester amniotic fluid (AF) from women with GDM. Targets of differentially expressed miRNAs were determined and pathway enrichment of target genes was performed. MiRNA and target gene expression were measured in a separate cohort of second trimester primary human fetal hepatocytes (PHFH) exposed to maternal obesity via qPCR and western blot. All studies were IRB approved.
Results
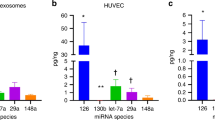
GDM-exposed AF had significant increases in miRNAs 199a-3p, 503-5p, and 1268a (fold change (FC) ≥ 1.5, p < 0.05). Female offspring-specific analysis showed enrichment in miRNAs 378a-3p, 885-5p, and 7-1-3p (p < 0.05). MiRNA gene targets were enriched in hepatic pathways. Key genes regulating de novo lipogenesis were upregulated in obesity-exposed PHFH, especially in males. Significantly altered miRNAs in GDM AF were measured in obese-exposed PHFH, with consistent increases in miRNAs 885-5p, 199-3p, 503-5p, 1268a, and 7-1-3p (FC ≥ 1.5, p < 0.05). Female PHFH exposed to maternal obesity had increased expression of miR-885-5p, miR-199-3p, miR-503-5p, miR-1268s, and miR-7-1-3p (p < 0.05), corresponding to decreased target genes expression for ABCA1, PAK4, and INSR. In male PHFHs, no miRNA changes were measured but there was increased expression of ABCA1, PAK4, and INSR (p < 0.05).
Conclusions
Our data suggest sex-specific changes in miRNA and gene expression in PHFH may be one mechanism contributing to the sexual dimorphism of metabolic disease in offspring exposed to GDM and maternal obesity in utero.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ashino NG, Saito KN, Souza FD, Nakutz FS, Roman EA, Velloso LA, et al. Maternal high-fat feeding through pregnancy and lactation predisposes mouse offspring to molecular insulin resistance and fatty liver. J Nutr Biochem. 2012;23:341–8.
Boney CM, Verma A, Tucker R, Vohr BR. Metabolic syndrome in childhood: association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics. 2005;115:e290–6.
Catalano PM, Ehrenberg HM. The short- and long-term implications of maternal obesity on the mother and her offspring. BJOG. 2006;113:1126–33.
Crume TL, Ogden L, West NA, Vehik KS, Scherzinger A, Daniels S, et al. Association of exposure to diabetes in utero with adiposity and fat distribution in a multiethnic population of youth: the Exploring Perinatal Outcomes among Children (EPOCH) Study. Diabetologia. 2011;54:87–92.
Kawasaki M, Arata N, Ogawa Y. Obesity and abnormal glucose tolerance in the offspring of mothers with diabetes. Curr Opin Obstet Gynecol. 2018;30:361–8.
McCurdy CE, Bishop JM, Williams SM, Grayson BE, Smith MS, Friedman JE, et al. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Investig. 2009;119:323–35.
Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM. Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia. 1993;36:62–7.
Plagemann A. Perinatal programming and functional teratogenesis: impact on body weight regulation and obesity. Physiol Behav. 2005;86:661–8.
Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118:2316–24.
Morales-Prieto DM, Ospina-Prieto S, Chaiwangyen W, Schoenleben M, Markert UR. Pregnancy-associated miRNA-clusters. J Reprod Immunol. 2013;97:51–61.
Mouillet JF, Chu T, Sadovsky Y. Expression patterns of placental microRNAs. Birth Defects Res A Clin Mol Teratol. 2011;91:737–43.
Mouillet JF, Ouyang Y, Coyne CB, Sadovsky Y. MicroRNAs in placental health and disease. Am J Obstet Gynecol. 2015;213(4 Suppl):S163–72.
Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41.
Underwood MA, Gilbert WM, Sherman MP. Amniotic fluid: not just fetal urine anymore. J Perinatol. 2005;25:341–8.
Pinney SE, Mesaros CA, Snyder NW, Busch CM, Xiao R, Aijaz S, et al. Second trimester amniotic fluid bisphenol A concentration is associated with decreased birth weight in term infants. Reprod Toxicol. 2017;67:1–9.
O'Neill K, Alexander J, Azuma R, Xiao R, Snyder NW, Mesaros CA, et al. Gestational diabetes alters the metabolomic profile in 2nd trimester amniotic fluid in a sex-specific manner. Int J Mol Sci. 2018;19:E2696.
Lazaro CA, Croager EJ, Mitchell C, Campbell JS, Yu C, Foraker J, et al. Establishment, characterization, and long-term maintenance of cultures of human fetal hepatocytes. Hepatology. 2003;38:1095–106.
Alejandro EU, Gregg B, Wallen T, Kumusoglu D, Meister D, Chen A, et al. Maternal diet-induced microRNAs and mTOR underlie beta cell dysfunction in offspring. J Clin Invest. 2014;124:4395–410.
Collares CV, Evangelista AF, Xavier DJ, Rassi DM, Arns T, Foss-Freitas MC, et al. Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients. BMC Res Notes. 2013;6:491.
Correa-Medina M, Bravo-Egana V, Rosero S, Ricordi C, Edlund H, Diez J, et al. MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas. Gene Expr Patterns. 2009;9:193–9.
Guay C, Regazzi R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol. 2013;9:513–21.
Hu L, Han J, Zheng F, Ma H, Chen J, Jiang Y, et al. Early second-trimester serum microRNAs as potential biomarker for nondiabetic macrosomia. Biomed Res Int. 2014;2014:394125.
Lazzarini R, Olivieri F, Ferretti C, Mattioli-Belmonte M, Di Primio R, Orciani M. mRNAs and miRNAs profiling of mesenchymal stem cells derived from amniotic fluid and skin: the double face of the coin. Cell Tissue Res. 2014;355:121–30.
Liu T, Chen Q, Huang Y, Huang Q, Jiang L, Guo L. Low microRNA-199a expression in human amniotic epithelial cell feeder layers maintains human-induced pluripotent stem cell pluripotency via increased leukemia inhibitory factor expression. Acta Biochim Biophys Sin (Shanghai). 2012;44:197–206.
Miranda-Sayago JM, Fernandez-Arcas N, Reyes-Engel A, Benito C, Narbona I, Alonso A. Changes in CDKN2D, TP53, and miR125a expression: potential role in the evaluation of human amniotic fluid-derived mesenchymal stromal cell fitness. Genes Cells. 2012;17:673–87.
Pillar N, Yoffe L, Hod M, Shomron N. The possible involvement of microRNAs in preeclampsia and gestational diabetes mellitus. Best Pract Res Clin Obstet Gynaecol. 2015;29:176–82.
Rayner KJ, Hennessy EJ. Extracellular communication via microRNA: lipid particles have a new message. J Lipid Res. 2013;54:1174–81.
Ross SA, Davis CD. The emerging role of microRNAs and nutrition in modulating health and disease. Annu Rev Nutr. 2014;34:305–36.
Tryggestad JB, Vishwanath A, Jiang S, Mallappa A, Teague AM, Takahashi Y, et al. Influence of gestational diabetes mellitus on human umbilical vein endothelial cell miRNA. Clin Sci (Lond). 2016;130:1955–67.
Vickers KC, Landstreet SR, Levin MG, Shoucri BM, Toth CL, Taylor RC, et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc Natl Acad Sci USA. 2014;111:14518–23.
Zhao C, Dong J, Jiang T, Shi Z, Yu B, Zhu Y, et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS ONE. 2011;6:e23925.
Zhao C, Zhang T, Shi Z, Ding H, Ling X. MicroRNA-518d regulates PPARalpha protein expression in the placentas of females with gestational diabetes mellitus. Mol Med Rep. 2014;9:2085–90.
Zhu Y, Tian F, Li H, Zhou Y, Lu J, Ge Q. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int J Gynaecol Obstet. 2015;130:49–53.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.
Qi R, Weiland M, Gao XH, Zhou L, Mi QS. Identification of endogenous normalizers for serum microRNAs by microarray profiling: U6 small nuclear RNA is not a reliable normalizer. Hepatology. 2012;55:1640–2.
Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005.
Stefulj J, Panzenboeck U, Becker T, Hirschmugl B, Schweinzer C, Lang I, et al. Human endothelial cells of the placental barrier efficiently deliver cholesterol to the fetal circulation via ABCA1 and ABCG1. Circ Res. 2009;104:600–8.
Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–43.
Lingwood BE, Henry AM, d'Emden MC, Fullerton AM, Mortimer RH, Colditz PB, et al. Determinants of body fat in infants of women with gestational diabetes mellitus differ with fetal sex. Diabetes Care. 2011;34:2581–5.
Regnault N, Gillman MW, Rifas-Shiman SL, Eggleston E, Oken E. Sex-specific associations of gestational glucose tolerance with childhood body composition. Diabetes Care. 2013;36:3045–53.
Retnakaran R, Kramer CK, Ye C, Kew S, Hanley AJ, Connelly PW, et al. Fetal sex and maternal risk of gestational diabetes mellitus: the impact of having a boy. Diabetes Care. 2015;38:844–51.
Alexander J, Teague AM, Chen J, Aston CE, Leung YK, Chernausek S, et al. Offspring sex impacts DNA methylation and gene expression in placentae from women with diabetes during pregnancy. PLoS ONE. 2018;13:e0190698.
He L, Tang M, Xiao T, Liu H, Liu W, Li G, et al. Obesity-associated miR-199a/214 cluster inhibits adipose browning via PRDM16-PGC-1alpha transcriptional network. Diabetes. 2018;67:2585–600.
Dimasuay KG, Boeuf P, Powell TL, Jansson T. Placental responses to changes in the maternal environment determine fetal growth. Front Physiol. 2016;7:12.
Jansson T, Aye IL, Goberdhan DC. The emerging role of mTORC1 signaling in placental nutrient-sensing. Placenta. 2012;33(Suppl 2):e23–9.
Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542–52.
Caporali A, Meloni M, Vollenkle C, Bonci D, Sala-Newby GB, Addis R, et al. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. 2011;123:282–91.
Shantikumar S, Caporali A, Emanueli C. Role of microRNAs in diabetes and its cardiovascular complications. Cardiovasc Res. 2012;93:583–93.
Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM, Hu RM. MicroRNA-320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clin Exp Pharmacol Physiol. 2009;36:181–8.
Zhou B, Ma R, Si W, Li S, Xu Y, Tu X, et al. MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett. 2013;333:159–69.
Carrer M, Liu N, Grueter CE, Williams AH, Frisard MI, Hulver MW, et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci USA. 2012;109:15330–5.
Huang N, Wang J, Xie W, Lyu Q, Wu J, He J, et al. MiR-378a-3p enhances adipogenesis by targeting mitogen-activated protein kinase 1. Biochem Biophys Res Commun. 2015;457:37–42.
Nieto M, Hevia P, Garcia E, Klein D, Alvarez-Cubela S, Bravo-Egana V, et al. Antisense miR-7 impairs insulin expression in developing pancreas and in cultured pancreatic buds. Cell Transplant. 2012;21:1761–74.
Filios SR, Shalev A. beta-Cell MicroRNAs: small but powerful. Diabetes. 2015;64:3631–44.
Kredo-Russo S, Mandelbaum AD, Ness A, Alon I, Lennox KA, Behlke MA, et al. Pancreas-enriched miRNA refines endocrine cell differentiation. Development. 2012;139:3021–31.
Raitoharju E, Seppala I, Lyytikainen LP, Viikari J, Ala-Korpela M, Soininen P, et al. Blood hsa-miR-122-5p and hsa-miR-885-5p levels associate with fatty liver and related lipoprotein metabolism-The Young Finns Study. Sci Rep. 2016;6:38262.
Hur K, Toiyama Y, Schetter AJ, Okugawa Y, Harris CC, Boland CR, et al. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J Natl Cancer Inst. 2015;107.
Mitsuhashi K, Yamamoto I, Kurihara H, Kanno S, Ito M, Igarashi H, et al. Analysis of the molecular features of rectal carcinoid tumors to identify new biomarkers that predict biological malignancy. Oncotarget. 2015;6:22114–25.
Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311:392–404.
Tan Y, Ge G, Pan T, Wen D, Gan J. Serum MiRNA panel as potential biomarkers for chronic hepatitis B with persistently normal alanine aminotransferase. Clin Chim Acta. 2015;451(Pt B):232–9.
Acknowledgements
We would like to thank Jeanne Manson, Ph.D and Deborah Driscoll, M.D. for their work in establishing the amniotic fluid and amniocyte biospecimen repository.
Funding
Research reported in this publication was supported by the National Institute of Environmental Health Sciences and the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under award numbers SEP: K08 DK090302, P30 ES013508, UL1TR001878, the McCabe Foundation and the creation of the biospecimen repository: 5R21-ES11675.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Joshi, A., Azuma, R., Akumuo, R. et al. Gestational diabetes and maternal obesity are associated with sex-specific changes in miRNA and target gene expression in the fetus. Int J Obes 44, 1497–1507 (2020). https://doi.org/10.1038/s41366-019-0485-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41366-019-0485-y
This article is cited by
-
Circulating mir-199-3p screens the onset of type 2 diabetes mellitus and the complication of coronary heart disease and predicts the occurrence of major adverse cardiovascular events
BMC Cardiovascular Disorders (2023)
-
MiR-17-5p/FOXL2/CDKN1B signal programming in oocytes mediates transgenerational inheritance of diminished ovarian reserve in female offspring rats induced by prenatal dexamethasone exposure
Cell Biology and Toxicology (2023)
-
Childhood obesity, metabolic syndrome, and oxidative stress: microRNAs go on stage
Reviews in Endocrine and Metabolic Disorders (2023)
-
The impact of fetal sex on risk factors for gestational diabetes and related adverse pregnancy outcomes
Acta Diabetologica (2022)