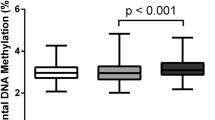
Abstract
Objectives
To study DNA methylation at the C19MC locus in the placenta and its association with (1) parental body size, (2) transmission of haplotypes for the C19MC rs55765443 SNP, and (3) offspring’s body size and/or body composition at birth and in childhood.
Subjects and methods
Seventy-two pregnant women-infant pairs and 63 fathers were included in the study. Weight and height of mothers, fathers and newborns were registered during pregnancy or at birth (n = 72). Placental DNA methylation at the C19MC imprinting control region (ICR) was quantified by bisulfite pyrosequencing. Genotyping of the SNP was performed using restriction fragment length polymorphisms. The children’s body size and composition were reassessed at age 6 years (n = 32).
Results
Lower levels of placental C19MC methylation were associated with increased body size of mother, specifically with higher pregestational and predelivery weights and height of the mother (β from –0.294 to –0.371; R2 from 0.04 to 0.10 and all p < 0.019), and with higher weight, height, waist and hip circumferences, and fat mass of the child (β from –0.428 to –0.552; R2 from 0.33 to 0.56 and all p < 0.009). Parental transmission of the SNP did not correlate with an altered placental methylation status at the C19MC ICR.
Conclusions
Increased maternal size is associated with reduced placental C19MC methylation, which, in turn, relate to larger body size of the child.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Koerner MV, Barlow DP. Genomic imprinting-an epigenetic gene-regulatory model. Curr Opin Genet Dev. 2010;20:164–70.
Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32.
Davies W, Isles AR, Humby T, Wilkinson LS. What are imprinted genes doing in the brain? Adv Exp Med Biol. 2008;626:62–70.
Tycko B. Imprinted genes in placental growth and obstetric disorders. Cytogenet Genome Res. 2006;113:271–8.
Smith FM, Garfield AS, Ward A. Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res. 2006;113:279–91.
Girardot M, Feil R, Lleres D. Epigenetic deregulation of genomic imprinting in humans: causal mechanisms and clinical implications. Epigenomics. 2013;5:715–28.
Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, et al. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet. 2010;19:3566–82.
Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70.
Bortolin-Cavaille ML, Dance M, Weber M, Cavaille J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37:3464–73.
Tsai KW, Kao HW, Chen HC, Chen SJ, Lin WC. Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics. 2009;4:587–92.
Mouillet JF, Ouyang Y, Coyne CB, Sadovsky Y. MicroRNAs in placental health and disease. Am J Obstet Gynecol. 2015;213:S163–172.
Hromadnikova I, Kotlabova K, Ondrackova M, Pirkova P, Kestlerova A, Novotna V, et al. Expression profile of C19MC microRNAs in placental tissue in pregnancy-related complications. DNA Cell Biol. 2015;34:437–57.
Xie L, Mouillet JF, Chu T, Parks WT, Sadovsky E, Knofler M, et al. C19MC microRNAs regulate the migration of human trophoblasts. Endocrinology. 2014;155:4975–85.
Ishida M, Moore GE. The role of imprinted genes in humans. Mol Aspects Med. 2013;34:826–40.
Richard N, Molin A, Coudray N, Rault-Guillaume P, Juppner H, Kottler ML. Paternal GNAS mutations lead to severe intrauterine growth retardation (IUGR) and provide evidence for a role of XLalphas in fetal development. J Clin Endocrinol Metabol. 2013;98:E1549–1556.
Bourque DK, Avila L, Penaherrera M, von Dadelszen P, Robinson WP. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not preeclampsia. Placenta. 2010;31:197–202.
Brehin AC, Colson C, Maupetit-Mehouas S, Grybek V, Richard N, Linglart A, et al. Loss of methylation at GNAS exon A/B is associated with increased intrauterine growth. J Clin Endocrinol Metabol. 2015;100:E623–631.
St-Pierre J, Hivert MF, Perron P, Poirier P, Guay SP, Brisson D, et al. IGF2 DNA methylation is a modulator of newborn’s fetal growth and development. Epigenetics. 2012;7:1125–32.
Prats-Puig A, Carreras-Badosa G, Bassols J, Cavelier P, Magret A, Sabench C, et al. The placental imprinted DLK1-DIO3 domain: a new link to prenatal and postnatal growth in humans. Am J Obstet Gynecol. 2017;217:350 e351–350 e313.
Grafodatskaya D, Cytrynbaum C, Weksberg R. The health risks of ART. EMBO Rep. 2013;14:129–35.
Carrascosa A, Fernandez JM, Fernandez C, Ferrandez A, Lopez-Siguero JP, Sanchez E, et al. Spanish growth studies 2008. New anthropometric standards. Endocrinol Nutr. 2008;55:484–506.
Treuth MS, Butte NF, Wong WW, Ellis KJ. Body composition in prepubertal girls: comparison of six methods. Int J Obes Relat Metabol Disord. 2001;25:1352–9.
Hirooka M, Kumagi T, Kurose K, Nakanishi S, Michitaka K, Matsuura B, et al. A technique for the measurement of visceral fat by ultrasonography: comparison of measurements by ultrasonography and computed tomography. Intern Med. 2005;44:794–9.
Ferrozzi F, Zuccoli G, Tognini G, Castriota-Scanderbeg A, Bacchini E, Bernasconi S, et al. An assessment of abdominal fatty tissue distribution in obese children. A comparison between echography and computed tomography. Radiol Med. 1999;98:490–4.
Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–75.
Carreras-Badosa G, Bonmati A, Ortega FJ, Mercader JM, Guindo-Martinez M, Torrents D, et al. Dysregulation of placental miRNA in maternal obesity is associated with pre- and postnatal growth. J Clin Endocrinol Metabol. 2017;102:2584–94.
Bellemer C, Bortolin-Cavaille ML, Schmidt U, Jensen SM, Kjems J, Bertrand E, et al. Microprocessor dynamics and interactions at endogenous imprinted C19MC microRNA genes. J Cell Sci. 2012;125:2709–20.
Donker RB, Mouillet JF, Chu T, Hubel CA, Stolz DB, Morelli AE, et al. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod. 2012;18:417–24.
Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet. 2014;15:517–30.
Martorell R, Zongrone A. Intergenerational influences on child growth and undernutrition. Paediatr Perinat Epidemiol. 2012;26:302–14.
Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–8.
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206.
Harvey NC, Poole JR, Javaid MK, Dennison EM, Robinson S, Inskip HM, et al. Parental determinants of neonatal body composition. J Clin Endocrinol Metabol. 2007;92:523–6.
Addo OY, Stein AD, Fall CH, Gigante DP, Guntupalli AM, Horta BL, et al. Maternal height and child growth patterns. J Pediatr. 2013;163:549–54.
Thame M, Osmond C, Trotman H. Fetal growth and birth size is associated with maternal anthropometry and body composition. Matern Child Nutr. 2015;11:574–82.
Borengasser SJ, Zhong Y, Kang P, Lindsey F, Ronis MJ, Badger TM, et al. Maternal obesity enhances white adipose tissue differentiation and alters genome-scale DNA methylation in male rat offspring. Endocrinology. 2013;154:4113–25.
Cooper WN, Khulan B, Owens S, Elks CE, Seidel V, Prentice AM, et al. DNA methylation profiling at imprinted loci after periconceptional micronutrient supplementation in humans: results of a pilot randomized controlled trial. FASEB J. 2012;26:1782–90.
Azzi S, Sas TC, Koudou Y, Le Bouc Y, Souberbielle JC, Dargent-Molina P, et al. Degree of methylation of ZAC1 (PLAGL1) is associated with prenatal and post-natal growth in healthy infants of the EDEN mother child cohort. Epigenetics. 2014;9:338–45.
Steegers-Theunissen RP, Obermann-Borst SA, Kremer D, Lindemans J, Siebel C, Steegers EA, et al. Periconceptional maternal folic acid use of 400 microg per day is related to increased methylation of the IGF2 gene in the very young child. PLoS ONE. 2009;4:e7845.
Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105:17046–9.
Cassidy FC, Charalambous M. Genomic imprinting, growth and maternal-fetal interactions. J Exp Biol. 2018;221(Pt Suppl 1) pii: jeb164517. https://doi.org/10.1242/jeb.164517.
Ivanova E, Chen JH, Segonds-Pichon A, Ozanne SE, Kelsey G. DNA methylation at differentially methylated regions of imprinted genes is resistant to developmental programming by maternal nutrition. Epigenetics. 2012;7:1200–10.
Radford EJ, Isganaitis E, Jimenez-Chillaron J, Schroeder J, Molla M, Andrews S, et al. An unbiased assessment of the role of imprinted genes in an intergenerational model of developmental programming. PLoS Genet. 2012;8:e1002605.
Kelsey G, Feil R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110336.
Meaburn EL, Schalkwyk LC, Mill J. Allele-specific methylation in the human genome: implications for genetic studies of complex disease. Epigenetics. 2010;5:578–82.
Tycko B. Allele-specific DNA methylation: beyond imprinting. Hum Mol Genet. 2010;19:R210–220.
Voisin S, Almen MS, Zheleznyakova GY, Lundberg L, Zarei S, Castillo S, et al. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome Med. 2015;7:103.
Chang G, Mouillet JF, Mishima T, Chu T, Sadovsky E, Coyne CB, et al. Expression and trafficking of placental microRNAs at the feto-maternal interface. FASEB J. 2017;31:2760–70.
Lin S, Cheung WK, Chen S, Lu G, Wang Z, Xie D, et al. Computational identification and characterization of primate-specific microRNAs in human genome. Comput Biol Chem. 2010;34:232–41.
Nguyen PN, Huang CJ, Sugii S, Cheong SK, Choo KB. Selective activation of miRNAs of the primate-specific chromosome 19 miRNA cluster (C19MC) in cancer and stem cells and possible contribution to regulation of apoptosis. J Biomed Sci. 2017;24:20.
Nguyen PNN, Choo KB, Huang CJ, Sugii S, Cheong SK, Kamarul T. miR-524-5p of the primate-specific C19MC miRNA cluster targets TP53IPN1- and EMT-associated genes to regulate cellular reprogramming. Stem Cell Res Ther. 2017;8:214.
Acknowledgements
The authors are grateful to all the children and parents who participated in the study. AP-P was granted with CAS14/00011 (Spanish Ministry of Education, Culture and Sport). SX-T holds a Sara Borrell contract from Carlos III National Institute of Health (ISCIII; CD15-00162). GC-B was granted a pFIS contract from ISCIII. BM-P holds a contract from Generalitat de Catalunya (SLT002/16/00065). JB is a Miguel Servet investigator from ISCIII (MS12/03239). FDZ is a Senior Investigator of the Clinical Research Fund of the Leuven University Hospital. LI is a Clinical Investigator of CIBERDEM (Spanish Biomedical Research Centre in Diabetes and Associated Metabolic Disorders) from ISCIII. AL-B is an I3 investigator (Spanish Ministry of Economy and Competitiveness). This study was supported by grants from the Ministerio de Ciencia e Innovación, Instituto de Salud Carlos III (ISCIII), Madrid, Spain (PI17/00557 to JB, and PI13/01257 and PI16/01335 to AL-B), projects co-funded by FEDER (Fondo Europeo de Desarrollo Regional). RF and MG acknowledge grant funding from the Fondation pour la Recherche Médicale (Equipes FRM, grant number DEQ31703).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Prats-Puig, A., Xargay-Torrent, S., Carreras-Badosa, G. et al. Methylation of the C19MC microRNA locus in the placenta: association with maternal and chilhood body size. Int J Obes 44, 13–22 (2020). https://doi.org/10.1038/s41366-019-0450-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41366-019-0450-9
This article is cited by
-
Cord blood epigenome-wide meta-analysis in six European-based child cohorts identifies signatures linked to rapid weight growth
BMC Medicine (2023)
-
Variation of microRNA expression in the human placenta driven by population identity and sex of the newborn
BMC Genomics (2021)
-
Maternal obesity alters C19MC microRNAs expression profile in fetal umbilical cord blood
Nutrition & Metabolism (2020)