Abstract
Background
Overweight and obesity can lead to adipose tissue inflammation, which causes insulin resistance and on the long-term type 2 diabetes mellitus (T2D). The inflammatory changes of obese-adipose tissue are characterized by macrophage infiltration and activation, but validated circulating biomarkers for adipose tissue inflammation for clinical use are still lacking. One of the most secreted enzymes by activated macrophages is chitotriosidase (CHIT1).
Objective
To test whether circulating CHIT1 enzymatic activity levels reflect adipose tissue inflammation.
Methods
Plasma and adipose tissue samples of 105 subjects (35 lean, 37 overweight, and 33 T2D patients) were investigated. CHIT1 mRNA levels were determined in adipose tissue-resident innate immune cells. CHIT1 mRNA levels, protein abundance, and plasma enzymatic activity were subsequently measured in adipose tissue biopsies and plasma of control subjects with varying levels of obesity and adipose tissue inflammation as well as in T2D patients.
Results
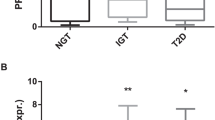
In adipose tissue, CHIT1 mRNA levels were higher in stromal vascular cells compared to adipocytes, and higher in adipose tissue-residing macrophages compared to circulating monocytes (p < 0.001). CHIT1 mRNA levels in adipose tissue were enhanced in overweightcompared to lean subjects and even more in T2D patients (p < 0.05). In contrast, plasma CHIT1 enzymatic activity did not differ between lean, overweight subjects and T2D patients. A mutation of the CHIT1 gene decreases plasma CHIT1 activity.
Conclusions
CHIT1 is expressed by adipose tissue macrophages and expression is higher in overweight subjects and T2D patients, indicating its potential as tissue biomarker for adipose tissue inflammation. However, these differences do not translate into different plasma CHIT1 activity levels. Moreover, a common CHIT1 gene mutation causing loss of plasma CHIT1 activity interferes with its use as a biomarker of adipose tissue inflammation. These results indicate that plasma CHIT1 activity is of limited value as a circulating biomarker for adipose tissue inflammation in human subjects.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Goossens GH. The role of adipose tissue dysfunction in the pathogenesis of obesity-related insulin resistance. Physiol Behav. 2008;94:206–18.
WHO. Global status report on noncommunicable diseases 2014. Geneva: World Health Organization; 2014.
Bluher M. Adipose tissue dysfunction contributes to obesity related metabolic diseases. Best Pract Res Clin Endocrinol Metab. 2013;27:163–77.
Trayhurn P. Hypoxia and adipose tissue function and dysfunction in obesity. Physiol Rev. 2013;93:1–21.
Tack CJ, Stienstra R, Joosten LA, Netea MG. Inflammation links excess fat to insulin resistance: the role of the interleukin-1 family. Immunol Rev. 2012;249:239–52.
Kusminski CM, Bickel PE, Scherer PE. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat Rev Drug Discov. 2016;15:639–60.
Odegaard JI, Chawla A. Type 2 responses at the interface between immunity and fat metabolism. Curr Opin Immunol. 2015;36:67–72.
Kohlgruber A, Lynch L. Adipose tissue inflammation in the pathogenesis of type 2 diabetes. Curr Diabetes Rep. 2015;15:92.
Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–808.
Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–30.
Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18:363–74.
Malaguarnera L. Chitotriosidase: the yin and yang. Cell Mol Life Sci. 2006;63:3018–29.
Sonmez A, Haymana C, Tapan S, Safer U, Celebi G, Ozturk O, et al. Chitotriosidase activity predicts endothelial dysfunction in type-2 diabetes mellitus. Endocrine. 2010;37:455–9.
Hollak CE, van Weely S, van Oers MH, Aerts JM. Marked elevation of plasma chitotriosidase activity. A novel hallmark of Gaucher disease. J Clin Invest. 1994;93:1288–92.
Kanneganti M, Kamba A, Mizoguchi E. Role of chitotriosidase (chitinase 1) under normal and disease conditions. J Epithel Biol Pharmacol. 2012;5:1–9.
Kabaroglu C, Onur E, Barutcuoglu B, Ozhan B, Erdinc S, Var A, et al. Inflammatory marker levels in obese adolescents with glucose intolerance: increased chitotriosidase activity. Clin Biochem. 2012;45:281–4.
Zurawska-Plaksej E, Knapik-Kordecka M, Rorbach-Dolata A, Piwowar A. Increased chitotriosidase activity in plasma of patients with type 2 diabetes. Arch Med Sci. 2016;12:977–84.
Zurawska-Plaksej E, Kratz EM, Ferens-Sieczkowska M, Knapik-Kordecka M, Piwowar A. Changes in glycosylation of human blood plasma chitotriosidase in patients with type 2 diabetes. Glycoconj J. 2016;33:29–39.
Zurawska-Plaksej E, Lugowska A, Hetmanczyk K, Knapik-Kordecka M, Adamiec R, Piwowar A. Proteins from the 18 glycosyl hydrolase family are associated with kidney dysfunction in patients with diabetes type 2. Biomarkers. 2015;20:52–7.
Ballak DB, van Diepen JA, Moschen AR, Jansen HJ, Hijmans A, Groenhof GJ, et al. IL-37 protects against obesity-induced inflammation and insulin resistance. Nat Commun. 2014;5:4711.
Ballak DB, van Asseldonk EJ, van Diepen JA, Jansen H, Hijmans A, Joosten LA, et al. TLR-3 is present in human adipocytes, but its signalling is not required for obesity-induced inflammation in adipose tissue in vivo. PLoS ONE. 2015;10:e0123152.
Jansen HJ, Stienstra R, van Diepen JA, Hijmans A, van der Laak JA, Vervoort GM, et al. Start of insulin therapy in patients with type 2 diabetes mellitus promotes the influx of macrophages into subcutaneous adipose tissue. Diabetologia. 2013;56:2573–81.
van Diepen JA, Robben JH, Hooiveld GJ, Carmone C, Alsady M, Boutens L, et al. SUCNR1-mediated chemotaxis of macrophages aggravates obesity-induced inflammation and diabetes. Diabetologia. 2017;60:1304–13.
Jackson AS, Pollock ML, Ward A. Generalized equations for predicting body density of women. Med Sci Sports Exerc. 1980;12:175–81.
Cinti S, Mitchell G, Barbatelli G, Murano I, Ceresi E, Faloia E, et al. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res. 2005;46:2347–55.
Lin K, Kools H, de Groot PJ, Gavai AK, Basnet RK, Cheng F, et al. MADMAX - management and analysis database for multiple ~omics experiments. J Integr Bioinforma. 2011;8:160.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64.
Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Sartor MA, Tomlinson CR, Wesselkamper SC, Sivaganesan S, Leikauf GD, Medvedovic M. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinforma. 2006;7:538.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol). 1995;57:289–300.
Moore DS, Notz, WI, Flinger, MA The basic practice of statistics. 6th ed. New York, NY: W. H. Freeman and Company; 2013.
Westerhuis JA, Hoefsloot HCJ, Smit S, Vis DJ, Smilde AK, van Velzen EJJ, et al. Assessment of PLSDA cross validation. Metabolomics. 2008;4:81–9.
Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst. 2001;58:109–30.
Cancello R, Henegar C, Viguerie N, Taleb S, Poitou C, Rouault C, et al. Reduction of macrophage infiltration and chemoattractant gene expression changes in white adipose tissue of morbidly obese subjects after surgery-induced weight loss. Diabetes. 2005;54:2277–86.
Sprenger R, Doneanu C, Langridge J, Vissers H, Aerts H. MRM quantification of chitotriosidase in human plasma. Application Note Waters Corporation.
Boot RG, Renkema GH, Verhoek M, Strijland A, Bliek J, de Meulemeester TM, et al. The human chitotriosidase gene. Nature of inherited enzyme deficiency. J Biol Chem. 1998;273:25680–5.
Di Rosa M, Malaguarnera G, De Gregorio C, D’Amico F, Mazzarino MC, Malaguarnera L. Modulation of chitotriosidase during macrophage differentiation. Cell Biochem Biophys. 2013;66:239–47.
Di Rosa M, De Gregorio C, Malaguarnera G, Tuttobene M, Biazzo F, Malaguarnera L. Evaluation of AMCase and CHIT-1 expression in monocyte macrophages lineage. Mol Cell Biochem. 2013;374:73–80.
Di Rosa M, Malaguarnera G, De Gregorio C, Drago F, Malaguarnera L. Evaluation of CHI3L-1 and CHIT-1 expression in differentiated and polarized macrophages. Inflammation. 2013;36:482–92.
Renkema GH, Boot RG, Strijland A, Donker-Koopman WE, van den Berg M, Muijsers AO, et al. Synthesis, sorting, and processing into distinct isoforms of human macrophage chitotriosidase. Eur J Biochem. 1997;244:279–85.
Malaguarnera L, Di Rosa M, Zambito AM, dell’Ombra N, Di Marco R, Malaguarnera M. Potential role of chitotriosidase gene in nonalcoholic fatty liver disease evolution. Am J Gastroenterol. 2006;101:2060–9.
Pecht T, Gutman-Tirosh A, Bashan N, Rudich A. Peripheral blood leucocyte subclasses as potential biomarkers of adipose tissue inflammation and obesity subphenotypes in humans. Obes Rev. 2014;15:322–37.
Eckel N, Li Y, Kuxhaus O, Stefan N, Hu FB, Schulze MB. Transition from metabolic healthy to unhealthy phenotypes and association with cardiovascular disease risk across BMI categories in 90 257 women (the Nurses’ Health Study): 30 year follow-up from a prospective cohort study. Lancet Diabetes Endocrinol. 2018;6:714–24.
Greenhill C. Redefining metabolically healthy obesity. Nat Rev Endocrinol. 2018. [Epub ahead of print].
McDonnell ME, Ganley-Leal LM, Mehta A, Bigornia SJ, Mott M, Rehman Q, et al. B lymphocytes in human subcutaneous adipose crown-like structures. Obesity . 2012;20:1372–8.
Curat CA, Wegner V, Sengenès C, Miranville A, Tonus C, Busse R, et al. Macrophages in human visceral adipose tissue: increased accumulation in obesity and a source of resistin and visfatin. Diabetologia. 2006;49:744.
Lee BC, Lee J. Cellular and molecular players in adipose tissue inflammation in the development of obesity-induced insulin resistance. Biochim Biophys Acta. 2014;1842:446–62.
Dahlman I, Kaaman M, Olsson T, Tan GD, Bickerton AS, Wahlen K, et al. A unique role of monocyte chemoattractant protein 1 among chemokines in adipose tissue of obese subjects. J Clin Endocrinol Metab. 2005;90:5834–40.
Murdolo G, Hammarstedt A, Sandqvist M, Schmelz M, Herder C, Smith U, et al. Monocyte chemoattractant protein-1 in subcutaneous abdominal adipose tissue: characterization of interstitial concentration and regulation of gene expression by insulin. J Clin Endocrinol Metab. 2007;92:2688–95.
Di Gregorio GB, Yao-Borengasser A, Rasouli N, Varma V, Lu T, Miles LM, et al. Expression of CD68 and macrophage chemoattractant protein-1 genes in human adipose and muscle tissues: association with cytokine expression, insulin resistance, and reduction by pioglitazone. Diabetes. 2005;54:2305–13.
Bruun JM, Lihn AS, Pedersen SB, Richelsen B. Monocyte chemoattractant protein-1 release is higher in visceral than subcutaneous human adipose tissue (AT): implication of macrophages resident in the AT. J Clin Endocrinol Metab. 2005;90:2282–9.
Kanda H, Tateya S, Tamori Y, Kotani K, Hiasa K-i, Kitazawa R, et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Investig. 2006;116:1494–505.
Martin BC, Warram JH, Krolewski AS, Soeldner JS, Kahn CR, Martin BC, et al. Role of glucose and insulin resistance in development of type 2 diabetes mellitus: results of a 25-year follow-up study. Lancet. 1992;340:925–9.
Elmonem MA, Amin HS, El-Essawy RA, Mehaney DA, Nabil M, Kamel LN, et al. Association of chitotriosidase enzyme activity and genotype with the risk of nephropathy in type 2 diabetes. Clin Biochem. 2016;49:444–8.
Kundak Ahmet A, Tascılar Mehmet E, Abaci A, Devrim I, Ozgen Ilker T, Demirpek U, et al. Serum chitotriosidase activity: is it a new inflammatory marker in obese children? J Pediatr Endocrinol Metab. 2012;25:63.
Turan E, Sozmen B, Eltutan M, Sozmen EY. Serum chitotriosidase enzyme activity is closely related to HbA1c levels and the complications in patients with diabetes mellitus type 2. Diabetes Metab Syndr. 2017;11:S503–S6.
Fusetti F, von Moeller H, Houston D, Rozeboom HJ, Dijkstra BW, Boot RG, et al. Structure of human chitotriosidase. Implications for specific inhibitor design and function of mammalian chitinase-like lectins. J Biol Chem. 2002;277:25537–44.
Di Rosa M, Malaguarnera L. Chitotriosidase: a new inflammatory marker in diabetic complications. Mol Cell Biol. 2016;83:211–9.
Malaguarnera L, Simpore J, Prodi DA, Angius A, Sassu A, Persico I, et al. A 24-bp duplication in exon 10 of human chitotriosidase gene from the sub-Saharan to the Mediterranean area: role of parasitic diseases and environmental conditions. Genes Immun. 2003;4:570–4.
Di Rosa M, Mangano K, De Gregorio C, Nicoletti F, Malaguarnera L. Association of chitotriosidase genotype with the development of non-alcoholic fatty liver disease. Hepatol Res. 2013;43:267–75.
Acknowledgements
This work is part of the Perspectief Biomarker Development Center Research Programme with project number 13543, which is (partly) financed by the Netherlands Organisation for Scientific Research (NWO). We thank Jacqueline Ratter for collecting and isolating CD14+ cells from adipose tissues and blood samples. We thank Astrid van Rens for measuring the plasma CHIT1 enzyme activity in blood plasma samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Tans, R., van Diepen, J.A., Bijlsma, S. et al. Evaluation of chitotriosidase as a biomarker for adipose tissue inflammation in overweight individuals and type 2 diabetic patients. Int J Obes 43, 1712–1723 (2019). https://doi.org/10.1038/s41366-018-0225-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41366-018-0225-8