Abstract
Background
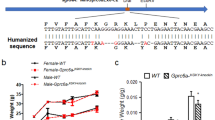
Replication initiator 1 (Repin1) is a zinc finger protein highly expressed in liver and adipose tissue. The Repin1 resides within a quantitative trait locus (QTL) for body weight and triglyceride levels in the rat, and its hepatic deletion in mice results in improved insulin sensitivity and lower body weight. Here, we analyzed whether genetic variation within the Repin1 affects parameters of glucose and lipid metabolism.
Methods
We sequenced REPIN1 in 48 non-related Caucasian subjects. We discovered a 12 base pair deletion (12 bp del; rs3832490), which was subsequently genotyped in two well-characterized cohorts (N = 3013) to test for associations with metabolic traits. Functional consequences of the variant were investigated in HepG2 cells in vitro.
Results
In human cohorts, we show that the 12 bp del associates with improved glucose metabolism (lower fasting plasma glucose, fasting plasma insulin, and HOMA IR). Cells transfected with the plasmid carrying the 12 bp del variant are characterized by increased GLUT2 and fatty acid translocase CD36 expression and more lipid droplets.
Conclusion
Our data suggest that genetic variation in human REPIN1 plays a role in glucose and lipid metabolism by differentially affecting the expression of REPIN1 target genes including glucose and fatty acid transporters.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206.
Kovács P, Klöting I. Quantitative trait loci on chromosomes 1 and 4 affect lipid phenotypes in the rat. Arch Biochem Biophys. 1998;354:139–43.
Klöting N, Wilke B, Klöting I. Triplet repeat in theRepin1 3′-untranslated region on rat chromosome 4 correlates with facets of the metabolic syndrome. Diabetes Metab Res Rev. 2007;23:406–10.
Caddle MS, Dailey L, Heintz NH. RIP60, a mammalian origin-binding protein, enhances DNA bending near the dihydrofolate reductase origin of replication. Mol Cell Biol. 1990;10:6236–6243.
Houchens CR, Montigny W, Zeltser L, Dailey L, Gilbert JM, Heintz NH. The dhfr oribeta-binding protein RIP60 contains 15 zinc fingers: DNA binding and looping by the central three fingers and an associated proline-rich region. Nucleic Acids Res. 2000;28:570–81.
Ruschke K, Illes M, Kern M, Klöting I, Fasshauer M, Schön MR, et al. Repin1 maybe involved in the regulation of cell size and glucose transport in adipocytes. Biochem Biophys Res Commun. 2010;400:246–51.
Kunath A, Hesselbarth N, Gericke M, Kern M, Dommel S, Kovacs P, et al. Repin1 deficiency improves insulin sensitivity and glucose metabolism in db/db mice by reducing adipose tissue mass and inflammation. Biochem Biophys Res Commun. 2016;478:398–402.
Hesselbarth N, Kunath A, Kern M, Gericke M, Mejhert N, Rydén M, et al. Repin1 deficiency in adipose tissue improves whole-body insulin sensitivity, and lipid metabolism. Int J Obes (Lond). 2017;41:1815–23.
Kern M, Kosacka J, Hesselbarth N, Bruckner J, Heiker JT, Flehmig G, et al. Liver-restricted repin1 deficiency improves whole-body insulin sensitivity, alters lipid metabolism, and causes secondary changes in adipose tissue in mice. Diabetes. 2014;63:3295–309.
Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806.
Standards of medical care in diabetes--2010. Diabetes Care American Diabetes Association 2010;33 (Suppl 1):S11–61.
Blüher M, Michael MD, Peroni OD, Ueki K, Carter N, Kahn BB, et al. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev Cell. 2002;3:25–38.
Tönjes A, Koriath M, Schleinitz D, Dietrich K, Böttcher Y, Rayner NW, et al. Genetic variation in GPR133 is associated with height: genome wide association study in the self-contained population of Sorbs. Hum Mol Genet. 2009;18:4662–8.
Tönjes A, Zeggini E, Kovacs P, Böttcher Y, Schleinitz D, Dietrich K, et al. Association of FTO variants with BMI and fat mass in the self-contained population of Sorbs in Germany. Eur J Hum Genet. 2010;18:104–10.
Veeramah KR, Tönjes A, Kovacs P, Gross A, Wegmann D, Geary P, et al. Genetic variation in the Sorbs of eastern Germany in the context of broader European genetic diversity. Eur J Hum Genet. 2011;19:995–1001.
Krowczynska AM, Coutts M, Makrides S, Brawerman G. The mouse homologue of the human acidic ribosomal phosphoprotein PO: a highly conserved polypeptide that is under translational control. Nucleic Acids Res. 1989;17:6408.
Mueckler M, Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. 2013;34:121–38.
Miquilena-Colina ME, Lima-Cabello E, Sanchez-Campos S, Garcia-Mediavilla MV, Fernandez-Bermejo M, Lozano-Rodriguez T, et al. Hepatic fatty acid translocase CD36 upregulation is associated with insulin resistance, hyperinsulinaemia and increased steatosis in non-alcoholic steatohepatitis and chronic hepatitis C. Gut. 2011;60:1394–402.
Krammer J, Digel M, Ehehalt F, Stremmel W, Fuellekrug J, Ehehalt R. Overexpression of CD36 and Acyl-CoA synthetases FATP2, FATP4 and ACSL1 increases fatty acid uptake in human hepatoma cells. Int J Med Sci. 2011;8:599–614.
Contreras AV, Torres N, Tovar AR. PPAR-alpha as a key nutritional and environmental sensor for metabolic adaptation. Adv Nutr. 2013;4:439–52.
Sato R. Sterol metabolism and SREBP activation. Arch Biochem Biophys. 2010;501:177–81.
White UA, Stephens JM. Transcriptional factors that promote formation of white adipose tissue. Mol Cell Endocrinol. 2010;318:10–4.
Diraison F, Parton L, Ferré P, Foufelle F, Briscoe CP, Leclerc I, et al. Over-expression of sterol-regulatory-element-binding protein-1c (SREBP1c) in rat pancreatic islets induces lipogenesis and decreases glucose-stimulated insulin release: modulation by 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR). Biochem J. 2004;378:769–78.
Borup A, Christensen PM, Nielsen LB, Christoffersen C. Apolipoprotein M in lipid metabolism and cardiometabolic diseases. Curr Opin Lipidol. 2015;26:48–55.
Xu N, Nilsson-Ehle P, Ahrén B. Correlation of apolipoprotein M with leptin and cholesterol in normal and obese subjects. J Nutr Biochem. 2004;15:579–82.
The Genotype-Tissue Expression (GTEx) project. GTEx Consortium Nat Genet. 2013;45:580–5.
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7.
Acknowledgements
We thank all those who participated in the studies. We would like to acknowledge excellent technical assistance by Ines Müller, Beate Gutsmann, Manuela Quandt, and Viola Döbel. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) (SFB 1052 “Obesity mechanisms’, projects B01, B03, B04, C01), from the Deutsche Diabetes Stiftung (DDS- Funktionelle Charakterisierung Adipositas- und Typ 2 Diabetes-assoziierter Varianten im Repin1-Gen), Deutsche Diabetes Gesellschaft (DDG 934300-003) and Deutsches Zentrum für Diabetesforschung (DZD 8200601). IFB Adiposity Diseases is supported by the Federal Ministry of Education and Research (BMBF), Germany, FKZ: 01EO1501 (AD2-060E, AD2-06E95, AD2-06E99). The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Krüger, J., Berger, C., Weidle, K. et al. Metabolic effects of genetic variation in the human REPIN1 gene. Int J Obes 43, 821–831 (2019). https://doi.org/10.1038/s41366-018-0123-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41366-018-0123-0
This article is cited by
-
Liver-specific Repin1 deficiency impairs transient hepatic steatosis in liver regeneration
Scientific Reports (2018)