Abstract
The first line of host defense against infectious agents involves activation of innate immune signaling pathways that recognize specific pathogen-associated molecular patterns (PAMPs). Key triggers of innate immune signaling are now known to include microbial-specific nucleic acid, which is rapidly detected in the cytosol of the cell. For example, RIG-I-like receptors (RLRs) have evolved to detect viral RNA species and to activate the production of host defense molecules and cytokines that stimulate adaptive immune responses. In addition, host defense countermeasures, including the production of type I interferons (IFNs), can also be triggered by microbial DNA from bacteria, viruses and perhaps parasites and are regulated by the cytosolic sensor, stimulator of interferon genes (STING). STING-dependent signaling is initiated by cyclic dinucleotides (CDNs) generated by intracellular bacteria following infection. CDNs can also be synthesized by a cellular synthase, cGAS, following interaction with invasive cytosolic self-DNA or microbial DNA species. The importance of STING signaling in host defense is evident since numerous pathogens have developed strategies to prevent STING function. Here, we review the relevance of STING-controlled innate immune signaling in host defense against pathogen invasion, including microbial endeavors to subvert this critical process.
Similar content being viewed by others
Introduction
The innate immune system comprises the foremost line of host defense to counter invasive microbial agents1,2. Over the past two decades, host pattern-recognition receptors (PRRs) have been shown to play a key role in recognizing non-self, pathogen-associated molecular patterns (PAMPs). A variety of PRRs have now been reported, including the Toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) and RIG-I-like receptors (RLRs)1. TLRs recognize extracellular or endosomal PAMPs, such as lipopolysaccharide (LPS), flagellin, single-stranded RNA, double-stranded RNA, and CpG DNA, to activate signaling through NF-κB, interferon regulatory factor (IRF) and MAP kinase signaling pathways, which induce cytokine production2. NLRs can also recognize PAMPs as well as damage-associated molecular patterns (DAMPs), including uric acid released by damaged cells, which trigger proinflammatory cytokine production3. The RLRs specifically recognize viral RNA species and activate analogous transcription factors and corresponding host defense-related molecules1. In addition, it is known that the presence of cytosolic DNA species can similarly trigger cytokine production4. This activity occurs because the cytosol is generally a DNA-free zone, and the existence of such nucleic acids usually signifies the arrival of an invading intracellular microbe or even leaked self-DNA from the nucleus as a result of DNA damage events. Cytosolic dsDNA species, generally over 70 bp in length, are now known to activate a host cyclic GMP-AMP synthase (cGAS), which generates cyclic dinucleotides (CDNs). These molecules bind, in turn, to an endoplasmic reticulum (ER)-associated sensor referred to as stimulator of interferon genes (STING), which results in NF-κB- and IRF3-dependent cytokine production4,5,6,7. Intracellular bacteria are also known to produce and secrete CDNs that directly activate STING signaling. Indeed, numerous DNA microbes have now been implicated in inadvertently triggering STING-dependent innate immune signaling and inducing cytokine production, including that of type I interferon (IFN)4,7. In response, there is growing evidence to indicate that a variety of microorganisms have attempted to evolve strategies to inhibit STING-dependent signaling. Here, we review the importance of STING-controlled innate immunity in preventing microbial infection, emphasizing how some of these pathogens try to subvert this critical host defense process. Understanding such host-pathogen interactions has important implications in the development of new therapeutic strategies to combat infectious disease.
Activation of STING signaling
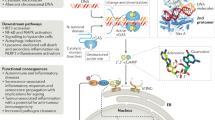
The sensor STING was discovered following high-throughput screening of cellular molecules that could activate the IFNβ promoter4,5. STING, also known as transmembrane protein 173 (TMEM173), is a 379 or 378 amino acid protein in human or mouse cells, respectively4,5,8,9,10. Under normal conditions, STING is localized in the ER and is expressed mainly in hematopoietic cells, including macrophages, dendritic cells, natural killer cells, and T cells, as well as in endothelial and epithelial cells, which might be exposed to the environment and thus susceptible to infectious agents4,5. STING is a sensor that is activated by CDNs, such as cyclic-di-AMP, cyclic-di-GMP, and cyclic-GMP-AMP (3′3′-cGAMP; cyclic[G(3′,5′)]pA(3′,5′)p), secreted by intracellular bacteria, such as Listeria monocytogenes, or by non-canonical cyclic-GMP-AMP (2′3′-cGAMP; cyclic[G(2′,5′)]pA(3′,5′)p)) generated by cGAS11,12,13,14,15,16,17. The sensing and interaction of CDNs induces a conformational change in STING and triggers the trafficking of STING complexed with TANK-binding kinase 1 (TBK1) from the ER to endosomal/lysosomal perinuclear regions4,5,18. This event mimics a form of autophagy4,19. Translocated TBK1 leads to phosphorylation of the transcription factors interferon regulatory factor 3 (IRF3) and nuclear factor-κB (NF-κB), which translocate to the nucleus and initiate innate immune gene transcription20,21 (Fig. 1). Following these events, STING activity is suppressed, and then STING is rapidly degraded to avoid sustained cytokine production, which could lead to autoinflammatory disease20.
STING is activated by cyclic dinucleotides (CDNs) secreted by intracellular bacteria or non-canonical CDNs generated by cGAS. The sensing and interaction of CDNs influences a conformational change in STING and triggers the trafficking of STING complexed with TBK1 from the ER to endosomal/lysosomal perinuclear regions. Translocated TBK1 leads to the phosphorylation of IRF3 and NF-kB to induce type I IFNs or inflammatory cytokines. Microbial DNA or RNA interacts with cGAS/STING to evade critical innate immune signaling. Red letters: DNA virus proteins, blue letters: RNA virus proteins
It is now well documented that STING plays an essential role in inducing type I IFN in response to sequence-nonspecific cytosolic DNA species that are greater than ~70 bp in human cells4,5. The requirement for large dsDNA species may be because cGAS needs to be in a dimeric form to be active, an event that requires two molecules of dsDNA, perhaps folded on themselves. Such DNA can constitute dsDNA oligonucleotides, single-stranded DNA forming hairpin duplexes, plasmids, and viral-, bacterial- or parasite-related DNA4,5. Sting knockout mice show high mortality following HSV-1 infection compared to that of wild-type mice4. STING has also been shown to be essential for the production of type I IFN induced by cytomegalovirus (CMV), vaccinia virus (VVΔE3L) and baculoviruses4. In addition, intracellular bacteria, such as Listeria monocytogenes and many others, may directly secrete STING-activating CDNs22,23. STING is not involved in dsRNA signaling, such as that by poly(I:C), which is largely governed by RLRs4. Nevertheless, loss of STING renders mice more susceptible to infection by select RNA viruses, such as vesicular stomatitis (VSV), suggesting that STING may play an important role in maintaining immune homeostasis4,5,24. Collectively, transient STING signaling plays a key role in protecting the host against a wide variety of pathogens, as described in more detail below. However, chronic STING activity may play a role in the development of autoinflammatory disease, underscoring the importance of tightly controlling this key innate immune signaling pathway25,26. This phenomenon may suggest that inflammatory events arising as a consequence of chronic infection may also involve the STING pathway, although this possibility remains to be clarified25.
DNA virus activation and evasion of STING-dependent innate immunity
A variety of DNA viruses have been reported to activate STING signaling4,5. The mechanisms remain unclear, but the majority of these viruses inject their genomes from their protective capsids into the nucleus when they reach the nuclear pore4,6,27,28. Thus, microbial DNA may be exposed and susceptible to interactions with cGAS/STING. STING or cGAS knockout mice, as well as isolated macrophages and dendritic cells from those mice, have been shown to be susceptible to herpes simplex virus 1 (HSV1) and other DNA viruses4,6. However, while such agents may inadvertently activate STING, many viruses have developed strategies to suppress STING signaling to survive. For example, a slew of HSV-encoded products, including ICP27, γ34.5, UL24, UL36, UL37, UL41, UL42, VP11/12, VP22, and VP24, have been reported to abrogate cGAS/STING-mediated signaling (Fig. 1 and Table 1)29,30,31,32,33,34,35,36,37. HSV encodes a large dsDNA genome of ~150,000 bp and predominantly remains in latency in peripheral neurons38. In one case, Christensen et al. showed that ICP27 translocated to the cytoplasm, where it interacted with TBK1 and STING and inhibited IRF3 activation29. HSV-1 γ34.5 has also been reported to inactivate STING through disrupting the trafficking of STING from the endoplasmic reticulum to the Golgi apparatus36. HSV 1 serine proteases VP22 and VP24 have been shown to selectively block STING agonist-induced phosphorylation and dimerization of IRF3 but not NF-κB activation30,31. VP22 also interacted with cGAS to inhibit its enzymatic activity31. UL24 was shown to prevent cGAS/STING-mediated IFNβ and interleukin-6 (IL-6) production by selectively blocking nuclear factor-κB (NF-κB) but not IFN-regulatory factor 3 function35. One of the most abundant HSV tegument proteins, UL46, was demonstrated to interact with STING to prevent activity32. It has also been reported that an additional tegument protein, UL41, reduced the accumulation of cGAS, which prevented CDN production33, and UL37 deamidated cGAS, similarly resulting in impaired CDN production34. Finally, HSV-1 ubiquitin-specific protease (UL36USP) antagonizes NF-kB activation induced by the STING pathway37. It is unclear why HSV may encode so many apparent ways to prevent STING signaling, but suppressing this pathway must be important for its survival. Perhaps this herpesvirus member utilizes varying suppressive methods at different stages of its life cycle, from entry to latency to its lytic phase.
However, another member of the herpesvirus family, Kaposi sarcoma herpes virus (KSHV), known as human herpesvirus 8 (HHV-8), is similarly a large double-stranded DNA virus able to trigger STING activity, which causes Kaposi’s sarcoma (KS)39. However, Ma et al. reported that KSHV-encoded vIRF1 inhibited this pathway by preventing STING from interacting with TBK140. Wu et al. additionally reported that KSHV ORF52, an abundant gamma herpesvirus-specific tegument protein, may impede CDN production through binding to both the DNA agonist and cGAS41. Furthermore, latency-associated nuclear antigen (LANA) of KSHV may inhibit STING signaling by directly binding to cGAS. This effect could conceivably antagonize cGAS-mediated restriction of KSHV’s lytic replication42. It remains to be seen whether other members of the herpesvirus family inhibit STING signaling. For example, CMV has been reported to trigger STING signaling following infection43,44. At least in mice, murine CMV (MCMV) may encode a product referred to as M152, which binds to STING to suppress this response45. Varicella zoster virus (VZV/HHV3) has also been documented to trigger STING signaling, although direct suppression of signaling has not yet been reported. It should be noted, however, that many of these and other viruses have also been shown to inhibit interferon signaling downstream of STING at the level of IRF3 or Jak/STAT signaling, indicating that suppression of host defense responses occurs at many levels46,47,48.
Hepatitis B virus (HBV), containing a circular DNA genome, specifically infects hepatocytes and causes chronic hepatitis49. Evidence indicates that HBV can decrease IFNβ production in transiently HBV-transfected Huh7 cells; stably HBV-producing cell lines, such as HepAD38; HBV-infected HepaRG cells; and primary human hepatocytes. The viral polymerase (Pol) of HBV has been reported to interfere with K63-linked polyubiquitination of STING via its reverse transcriptase (RT) domain50. However, it is still controversial whether HBV infection elicits a detectable cytokine response in hepatocytes, at least through STING. While one group reported that human hepatoma cells as well as immortalized mouse hepatocytes express low levels of STING51,52, another group indicated that human and murine hepatocytes do not express STING and do not produce type I IFN in response to foreign DNA or HBV infection51. Indeed, it is tempting to speculate that some viruses may target cells that may lack certain innate immune sensing pathways. Nevertheless, Kupffer cells, as stellate macrophages located in the liver, may express STING, and contribute toward the clearance of dying infected hepatocytes to possibly influence inflammation.
Other double-stranded DNA viruses, such as adenovirus (Ad) and human papillomavirus (HPV), have similarly been shown to antagonize the cGAS/STING DNA-sensing pathway27,28,53. Following Ad infection, cells deficient in STING or cGAS expression were noted to lack IRF3 phosphorylation, and activation of IFNβ or IRF3-responsive genes, such as ISG15 and ISG54, was compromised27,53. The oncogene E1A from Ad and E7 from HPV reportedly inhibit the cGAS/STING pathway by directly binding to STING. Suppression of E1A and E7 expression could restore the production of type I IFNs28. Finally, Eaglesham et al. showed that the large cytosolic DNA virus, vaccinia virus similarly suppresses STING via the production of poxins which cleave CDNs. Collectively, it is perhaps unsurprising that DNA viruses have evolved mechanisms to suppress dsDNA-triggered innate immune signaling. Many of the viruses noted here can remain latent and even contribute toward tumorigenesis. It is unclear whether suppression of STING signaling may influence the transformation process. Evidence now indicates that STING signaling is suppressed in many types of tumor cells, presumably to avoid DNA damage-activated immune responses54,55. In addition, STING activity has been shown to be important for the generation of antiviral as well as antitumor T cells. Thus, suppression of cGAS/STING not only may help DNA viruses survive but also may contribute toward cellular transformation.
The STING signaling pathway and retroviral infection
Host defense gene induction has been reported to also occur following retrovirus/lentivirus entry56. Following infection, the viral single-stranded RNA genome is reverse transcribed and delivered to the nucleus via mature integration complexes. STING has been reported to colocalize with such complexes57. Perhaps as a result, cGAS/STING knockout mice are defective in HIV-, murine leukemia virus-, and simian immunodeficiency virus-triggered type I IFN production7. However, the production of type I IFN is generally weak. This phenomenon may be due to agonist viral DNA species in the cytosol being degraded by cytoplasmic DNases, such as Trex1, a 3′–5′ exonuclease58. In the absence of Trex1, genomic or viral DNA accumulates in the cytosol and activates STING-dependent innate immune signaling59. In humans, mutations in Trex1 cause inflammatory diseases, such as Aicardi-Goutieres syndrome (AGS) and severe systemic lupus erythematosus (SLE)26,60,61. In experimental conditions, Trex1 deficiency reportedly results in increased HIV replication and type I IFN production61. Moreover, two single nucleotide polymorphisms (SNPs) in Trex1 have been documented in humans as being associated with faster HIV-1 disease progression and increased HIV replication62. Another negative regulator of innate immunity, a member of the nucleotide-binding domain, leucine-rich repeat-containing proteins (NLRs), NLRX1, has also been described as associating with STING to reduce TBK1 activity and enable increased HIV-1 infection63,64. Human T lymphotropic virus type 1 (HTLV-1), a member of the delta retrovirus family, is the causative agent of adult T cell leukemia (ATL) and tropical spastic paraparesis (TSP)65. HTLV-1 reverse transcription intermediates (RTIs) have been shown to trigger STING-dependent IFNβ production in differentiated human macrophages, including THP1 cells. It has also been reported that HTLV-1 RTIs interact with STING and induce IRF3-Bax complexation, leading to apoptosis66. The HTLV-1 protein Tax has been shown to impair IFNβ production by influencing K63-linked ubiquitination of STING to disrupt interactions between STING and TANK-binding kinase 1 (TBK1)67. Thus, retroviruses/lentiviruses have evolved to avoid robust STING activation and may be assisted by molecules such as Trex1. It should be noted that up to 10% of the human genome contains versions of ancient retroviruses referred to as human endogenous retroviruses (ERVs)68. In addition, over 40% of the human genome consists of retrotransposons, which are DNA components that can be transcribed into RNA and converted back into identical DNA sequences by a reverse transcriptase encoded by the retrotransposon itself68. It is unclear whether such ERVs or retrotransposons aggravate innate immune signaling when reactivated to cause inflammatory disease59,69,70.
RNA virus infection and STING-dependent innate immunity
As discussed, STING signaling controls CDN- and cytosolic DNA-triggered innate immune signaling. However, early studies quickly showed that STING knockout mice were also susceptible to RNA viruses, such as VSV4. Usually, these pathways are governed by the RLR pathway, TLR3 and TLR71,2. However, type I IFN production was noted as being decreased in STING knockout cells infected with VSV. This result implies that STING is also necessary for protection against certain RNA viruses4. Recently, it was reported that STING may also restrict the replication of various RNA viruses at the posttranslational level71. This effect may be due to STING residing in the ER of the cell and being associated with the translocon, a portal where proteins destined for glycosylation and/or secretion are held for appropriate maturation5,25. The role of STING in translocon function remains to be clarified. Regardless, growing evidence now indicates that certain RNA viruses target STING for suppression (Fig. 1 and Table 2).
Hepatitis C virus (HCV) is an enveloped, positive-sense, single-stranded RNA virus in the family Flaviviridae that causes hepatitis and facilitates cancer development, such as that of hepatocellular carcinoma72. NS3/4A and NS4B, a serine protease of HCV, targets IPS1/MAVS/Cardif, a CARD-containing adaptor protein to block type I IFN production via RLRs73. In addition, STING-dependent IFNβ activation was observed to be suppressed by NS4B74. It is possible that NS4B disrupts the interaction between STING and TBK in STING- and TBK1-overexpressing cells transfected with the NS4B plasmid75,76. Similar to the situation with HBV infection, it is not clear whether STING is highly expressed in HCV-infected hepatocytes. However, Kupffer cells may play a key role in viral clearance and plausibly in inflammation associated with hepatitis-related diseases.
Dengue virus (DENV) is a mosquito-borne single-, positive-stranded RNA virus belonging to Flaviviridae that causes hemorrhagic fever in humans77. It has been documented that the DENV NS2B3 protease can inhibit type I IFN production through its proteolytic activity. It was shown that the protease of DENV targets and cleaves wildtype STING to prevent type I IFN production. DENV replication is highly increased in STING-deficient primary cells78,79. Recently, Aguirre et al. reported that NS2B also targets cGAS for degradation in an autophagy-lysosome-dependent mechanism to prevent sensing of mitochondrial DNA released during DENV infection80. Furthermore, the protease of dengue virus 2 (DENV2) cleaves human but not primate STING, reducing type I interferon production and boosting viral titers81. However, another positive-stranded RNA virus, which closely resembles DENV, is Zika virus (ZIKV), first isolated in Uganda in 1947. Recently, a large outbreak of malaise was identified as involving Zika infection in Brazil in 2015; thereafter, cases of outbreaks and evidence of transmission soon appeared worldwide, including in the Americas. It has been reported that different non-structural proteins of ZIKV, such as NS1 and NS4B, decrease the innate antiviral response to evade the host immune response82. Similar to DENV, the NS2B3 protease of ZIKV cleaves R78 and G79 in the cytoplasmic loop of human STING83. In an analysis of the host tropism of ZIKV, rodents, unlike humans, are not susceptible to ZIKV infection. This difference may be due to R78 and G79 being only partially conserved in the murine ortholog of STING83. In addition, Zheng et al. have shown that the NS1 protein of ZIKV recruits the deubiquitinase USP8 to cleave K11-linked ubiquitin chains at lysine 134 of caspase-1. Subsequently, caspase-1 targets cGAS for cleavage, which results in a reduction in type IFN production84. ZIKV is known to cause microcephaly in newborns, although the mechanisms and frequency of this syndrome remain to be clarified. One group has shown that STING-dependent signaling plays a role in antiviral macroautophagy/autophagy to restrict ZIKV infection in the fly brain. This study in Drosophila reveals key insights into the evolutionary function of STING in antiviral defense and further evidence for the ancestral function of autophagy in protecting host cells from viral invaders85,86,87.
Influenza A viruses (IAVs), in contrast, are negative-sense, single-stranded, segmented viruses that may suppress STING signaling88. In this regard, the hemagglutinin fusion peptide (FP) of IAV reportedly interacts with STING to antagonize type I IFN production in a STING-dependent but cGAS-independent manner89.
Thus, STING may also play an evolutionarily important role in protecting the host against microbial infection. In this light, it is worth noting that many RNA viruses, including DENV and ZIKV, are able to infect both human and insect cells. It is unclear whether such viruses suppress STING in their insect hosts if STING is expressed. Indeed, many viruses may only be able to succeed in hosts/cells where STING or similar innate immune pathways are absent.
Bacteria, CDNs, and STING-dependent innate immunity
STING is a direct sensor of CDNs, including c-di-GMP and c-di-AMP, generated by numerous intracellular bacteria, such as Listeria monocytogenes90. CDNs play a significant role in the life cycle of such bacteria, functioning as second messengers11. Listeria monocytogenes (L. monocytogenes) infection reportedly induces type I IFN and IL6 in wild-type murine fibroblasts, macrophages, and dendritic cells and in vivo, which is dependent on STING via CDNs4,23,91. L. monocytogenes secretes c-di-AMP through multidrug efflux pumps (MEPs)11. Moreover, L. monocytogenes DNA is also able to stimulate the IFN response in the STING/cGAS pathway in human macrophages22. STING likely evolved to detect CDNs early in evolution. The synthase cGAS probably later evolved to generate CDNs following interaction with DNA. Thus, STING may have been predominantly involved in innate immunity to bacterial infection and even RNA virus infection (through its speculative translocon function) before becoming central in innate immune signaling pathways triggered by DNA5.
Extracellular pathogens, such as Streptococcus pneumoniae (S. pneumoniae), are some of the leading causes of death in people over the age of 65 years. S. pneumoniae has been known to induce type I IFN and to regulate RANTES production through STING92,93. STING-dependent type I IFN production in elderly mice was decreased following S. pneumoniae infection. S. pneumoniae infection induces ER stress and augments inositol-requiring protein 1/X-box binding protein 1-mediated production of autophagy-related gene 9 (Atg9a)94. Saito et al. showed that a loss of Atg9 enhances the assembly of STING/TBK1 and increases innate immune signaling19. This result indicates that Atg9 induction by ER stress could decrease STING activity by S. pneumoniae infection, providing new evidence as to why older people may be more susceptible to infection.
Mycobacteria tuberculosis (M. tuberculosis), the causative agent of tuberculosis, remains one of the leading causes of chronic infectious pulmonary disease95. M. tuberculosis activates a cytosolic surveillance pathway (CSP) and induces innate immune responses following perforation of the phagosome membrane. This effect is mediated by the microbe’s ESX-1 secretion system following interaction with target macrophages96. Permeabilization mediated by ESX-1 allows cytosolic components of the ubiquitin-mediated autophagy pathway access to M. tuberculosis in phagosomes. Consequently, the STING pathway recognizes the extracellular bacterial DNA and activates innate immune responses96,97,98,99. CDNs can also be generated by such microbes, which can directly activate STING100,101. Dey et al. reported that c-di-AMP produced by M. tuberculosis controls the fate of infection by stimulating IFNβ production, an event that may actually facilitate bacterial survival100,101.
In addition to the bacteria described here, various other microbes, such as Chlamydia, Francisella, Brucella, Shigella, Salmonella, and Neisseria, have been reported to engage the STING-dependent pathway102. However, while STING has likely evolved to recognize bacterial infection through recognition of the CDNs produced, the role of such CDNs in manipulating STING signaling, perhaps even to facilitate their survival, remains an interesting area of study, which will likely help explain mechanisms of pathogenesis102.
Parasites, malaria and STING signaling
Plasmodium parasites cause malaria, a debilitating disease affecting millions worldwide. Malaria infection is initiated by mosquitos injecting infectious sporozoites following biting their host. These sporozoites are transferred to the liver via the bloodstream. After replication in the liver, infectious exoerythrocytic merozoites are released into the blood103. Miller et al. showed that plasmodiums in the liver induce type I interferon-mediated innate immune responses. Type I IFN activates NKT cells, which produce IFNγ to inhibit secondary liver-stage infection104. Malaria-specific parasites inside red blood cells secrete extracellular vesicles (EVs) containing parasitic small RNA and genomic DNA. Human monocytes can take up the EVs, and parasitic DNA is released into the host cell cytosol, where STING is activated105. However, it has also been shown that TLR7 in pDCs can also contribute to type I IFN production in response to malaria infection in a murine model106. Thus, STING signaling may contribute toward protection of the host against malaria. Whether STING also plays a crucial role in protecting the host against other types of parasites remains to be seen.
Potential of STING in new antipathogen strategies
STING signaling plays an important role in stimulating the immune system in response to microbial infection, suggesting that control of this pathway may be useful in antimicrobial strategies to control disease. As described, various CDNs, such as cyclic-di-AMP, cyclic-di-GMP, and cGAMP (and synthetic analogues), can stimulate STING activity12,14,15,16. Indeed, STING agonists are now being evaluated in the clinic to enhance antitumor immunity107,108,109,110. Evidence indicates that the injection of CDNs into tumors stimulates surrounding antigen-presenting cells (APCs) to augment antitumor CTL activity111,112. Similarly, it is possible that comparable strategies may exert useful antimicrobial activity. In one example, reports indicate that systemic or local application of 2′3′-cGAMP reduces genital HSV-2 replication and improves the clinical outcome of infection, with strong induction of type I IFNs both in human cells and in mice in vivo113.
In addition to CDNs, alternate STING agonists have also been reported. For example, 5,6-dimethylxanthenone-4-acetic acid (DMXAA) and 10-(carboxymethyl)-9(10H) acridone (CMA) are flavonoids that potently bind to and activate STING signaling114,115. In a hepatitis B virus (HBV) hydrodynamic mouse model, DMXAA induced IFN-stimulated genes and decreased HBV DNA replication in the livers of mice. Since chronic HBV infection involves failure of the host to induce a sufficient immune response to clear the virus, such strategies indicate that activation of the STING pathway by agonists may be useful in treating such diseases116. In another example, a group identified novel IFN/IRF3-inducing molecules by high-throughput in vitro screening, referred to as 4-(2-chloro-6-fluorobenzyl)-N-(furan-2-ylmethyl)-3-oxo-3,4-dihydro-2H-benzo[b] thiazine-6-carboxamide (G10), and N-(methylcarbamoyl)-2-{[5-(4-methylphenyl)-1,3,4-oxadiazol-2-yl]sulfanyl}-2-phenylacetamide (C11)117,118. G10 reportedly induced IFN/IRF3-dependent signaling but not NFĸB signaling. This compound mediated anti alphaviral activity against chikungunya virus (CHIKV), Venezuelan equine encephalitis virus (VEEV) and Sindbis virus (SINV) and required STING- but not IPS-1/MAVS-dependent signaling118. C11 was also able to induce IFN secretion in human cells in a manner that required STING but not MAVS or TRIF. C11-treated cells potently blocked the replication of multiple emerging alphavirus types, including chikungunya, Ross River, Venezuelan equine encephalitis, Mayaro, and O’nyong’nyong viruses117. Thus, the use of STING agonists may be of benefit in treating microbial disease as well as in immune cancer therapy.
Finally, it is noteworthy that STING agonists may also be useful as vaccine adjuvants for the stimulation of the STING-dependent innate immune pathway. A number of examples now demonstrate the usefulness of such approaches in vaccine development to protect against microbes107,108,109,110,116,117,118. For example, CDN-formulated vaccines elicited long-lasting protective immunity against Mycobacterium tuberculosis in a murine model similar to that elicited by live attenuated vaccine strains presently in use, such as Bacille Calmette-Guérin (BCG)119.
The discovery of the STING signaling pathway has provided considerable insight into microbial pathogenesis, mechanisms of host defense, and causes of inflammatory disease and even cancer. These discoveries have led to investigation of whether controlling the STING pathway can generate new vaccines as well as antimicrobial agents to control a variety of diseases.
References
Kumar, H., Kawai, T. & Akira, S. Pathogen recognition by the innate immune system. Int Rev. Immunol. 30, 16–34 (2011).
Kawai, T. & Akira, S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 (2010).
Chen, G. Y. & Nunez, G. Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol. 10, 826–837 (2010).
Ishikawa, H., Ma, Z. & Barber, G. N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009).
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678, https://doi.org/10.1038/nature07317 (2008).
Li, X. D. et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394 (2013).
Gao, D. et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341, 903–906 (2013).
Jin, L. et al. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 28, 5014–5026 (2008).
Sun, W. et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl Acad. Sci. USA 106, 8653–8658 (2009).
Zhong, B. et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008).
Woodward, J. J., Iavarone, A. T. & Portnoy, D. A. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705 (2010).
Wu, J. et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013). science.1229963 [pii].
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Burdette, D. L. et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011).
Ablasser, A. et al. cGAS produces a 2’-5’-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 (2013).
Danilchanka, O. & Mekalanos, J. J. Cyclic dinucleotides and the innate immune response. Cell 154, 962–970 (2013).
Shang, G., Zhang, C., Chen, Z. J., Bai, X. C. & Zhang, X. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 567, 389–393 (2019).
Zhang, C. et al. Structural basis of STING binding with and phosphorylation by TBK1. Nature 567, 394–398 (2019).
Saitoh, T. et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl Acad. Sci. USA 106, 20842–20846 (2009).
Konno, H., Konno, K. & Barber, G. N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155, 688–698 (2013).
Tanaka, Y. & Chen, Z. J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal 5, ra20 (2012).
Hansen, K. et al. Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 33, 1654–1666 (2014).
Sauer, J. D. et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79, 688–694 (2011).
Ahn, J., Son, S., Oliveira, S. C. & Barber, G. N. STING-dependent signaling underlies IL-10 controlled inflammatory colitis. Cell Rep. 21, 3873–3884 (2017).
Barber, G. N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 15, 760–770 (2015).
Ahn, J. & Barber, G. N. Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Curr. Opin. Immunol. 31, 121–126 (2014).
Lam, E., Stein, S. & Falck-Pedersen, E. Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J. Virol. 88, 974–981 (2014).
Lau, L., Gray, E. E., Brunette, R. L. & Stetson, D. B. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 350, 568–571 (2015).
Christensen, M. H. et al. HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J. 35, 1385–1399 (2016).
Zhang, D., Su, C. & Zheng, C. Herpes simplex virus 1 serine protease VP24 Blocks the DNA-sensing signal pathway by abrogating activation of interferon regulatory factor 3. J. Virol. 90, 5824–5829 (2016).
Huang, J. et al. Herpes simplex virus 1 tegument protein VP22 abrogates cGAS/STING-mediated antiviral innate immunity. J. Virol 92, https://doi.org/10.1128/JVI.00841-18 (2018).
Deschamps, T. & Kalamvoki, M. Evasion of the STING DNA-sensing pathway by VP11/12 of herpes simplex virus 1. J. Virol 91, https://doi.org/10.1128/JVI.00535-17 (2017).
Su, C. & Zheng, C. Herpes simplex virus 1 abrogates the cGAS/STING-mediated cytosolic DNA-sensing pathway via its virion host shutoff protein, UL41. J Virol 91, https://doi.org/10.1128/JVI.02414-16 (2017).
Zhang, J. et al. Species-specific deamidation of cGAS by herpes simplex virus UL37 protein facilitates viral replication. Cell Host Microbe 24, 234–248 e235 (2018).
Xu, H., Su, C., Pearson, A., Mody, C.H. & Zheng, C. Herpes simplex virus 1 UL24 abrogates the DNA sensing signal pathway by inhibiting NF-kappaB activation. J. Virol. 91, https://doi.org/10.1128/JVI.00025-17 (2017).
Pan, S., Liu, X., Ma, Y., Cao, Y. & He, B. Herpes simplex virus 1 gamma134.5 protein inhibits STING activation that restricts viral replication. J. Virol. 92, https://doi.org/10.1128/JVI.01015-18 (2018).
Ye, R., Su, C., Xu, H. & Zheng, C. Herpes simplex virus 1 ubiquitin-specific protease UL36 abrogates NF-kappaB activation in DNA sensing signal pathway. J. Virol. 91, https://doi.org/10.1128/JVI.02417-16 (2017).
Whitley, R. J., Kimberlin, D. W. & Roizman, B. Herpes simplex viruses. Clin. Infect. Dis. 26, 541–553 (1998). quiz 554-545.
Horan, K. A. et al. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J. Immunol. 190, 2311–2319 (2013).
Ma, Z. et al. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc. Natl Acad. Sci. USA 112, E4306–E4315 (2015).
Wu, J. J. et al. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe 18, 333–344 (2015).
Zhang, G. et al. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc. Natl. Acad. Sci. USA 113, E1034–E1043 (2016).
Bianco, C. & Mohr, I. Restriction of human cytomegalovirus replication by ISG15, a host effector regulated by cGAS-STING double-stranded-DNA sensing. J. Virol. 91, https://doi.org/10.1128/JVI.02483-16 (2017).
Lio, C. W. et al. cGAS-STING signaling regulates initial innate control of cytomegalovirus infection. J. Virol. 90, 7789–7797 (2016).
Stempel, M. et al. The herpesviral antagonist m152 reveals differential activation of STING-dependent IRF and NF-kappaB signaling and STING’s dual role during MCMV infection. EMBO J 38, https://doi.org/10.15252/embj.2018100983 (2019).
Kim, J. A., Park, S. K., Kumar, M., Lee, C. H. & Shin, O. S. Insights into the role of immunosenescence during varicella zoster virus infection (shingles) in the aging cell model. Oncotarget 6, 35324–35343 (2015).
Kim, J. A., Park, S. K., Seo, S. W., Lee, C. H. & Shin, O. S. STING is involved in antiviral immune response against VZV Infection via the Induction of type I and III IFN pathways. J. Invest Dermatol 137, 2101–2109 (2017).
Gershon, M. & Gershon, A. Varicella-zoster virus and the enteric nervous system. J. Infect. Dis. 218, S113–S119 (2018).
Farazi, P. A. & DePinho, R. A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer 6, 674–687 (2006).
Liu, Y. et al. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J. Virol. 89, 2287–2300 (2015).
Thomsen, M. K. et al. Lack of immunological DNA sensing in hepatocytes facilitates hepatitis B virus infection. Hepatology 64, 746–759 (2016).
Guo, F. et al. Activation of stimulator of interferon genes in hepatocytes suppresses the replication of hepatitis B virus. Antimicrob Agents Chemother 61, https://doi.org/10.1128/AAC.00771-17 (2017).
Anghelina, D., Lam, E. & Falck-Pedersen, E. Diminished innate antiviral response to adenovirus vectors in cGAS/STING-deficient mice minimally impacts adaptive immunity. J. Virol. 90, 5915–5927 (2016).
Xia, T., Konno, H., Ahn, J. & Barber, G. N. Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 14, 282–297 (2016).
Xia, T., Konno, H. & Barber, G. N. Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis. Cancer Res 76, 6747–6759 (2016).
Doyle, T., Goujon, C. & Malim, M. H. HIV-1 and interferons: who’s interfering with whom? Nat. Rev. Microbiol 13, 403–413 (2015).
Jakobsen, M. R. et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc. Natl Acad. Sci. USA 110, E4571–E4580 (2013).
Yan, N., Cherepanov, P., Daigle, J. E., Engelman, A. & Lieberman, J. The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog. 5, e1000327 (2009).
Ahn, J. et al. Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 5, 5166 (2014).
Lee-Kirsch, M. A. et al. Mutations in the gene encoding the 3’-5’ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 39, 1065–1067 (2007).
Yan, N., Regalado-Magdos, A. D., Stiggelbout, B., Lee-Kirsch, M. A. & Lieberman, J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 11, 1005–1013 (2010).
Booiman, T., Setiawan, L. C. & Kootstra, N. A. Genetic variation in Trex1 affects HIV-1 disease progression. AIDS 28, 2517–2521 (2014).
Guo, H. et al. NLRX1 sequesters STING to negatively regulate the interferon response, thereby facilitating the replication of HIV-1 and DNA Viruses. Cell Host Microbe 19, 515–528 (2016).
Allen, I. C. et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity 34, 854–865 (2011).
Ishitsuka, K. & Tamura, K. Human T-cell leukaemia virus type I and adult T-cell leukaemia-lymphoma. Lancet Oncol. 15, e517–e526 (2014).
Sze, A. et al. Host restriction factor SAMHD1 limits human T cell leukemia virus type 1 infection of monocytes via STING-mediated apoptosis. Cell Host Microbe 14, 422–434 (2013).
Wang, J., Yang, S., Liu, L., Wang, H. & Yang, B. HTLV-1 Tax impairs K63-linked ubiquitination of STING to evade host innate immunity. Virus Res 232, 13–21 (2017).
Grandi, N. & Tramontano, E. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front Immunol. 9, 2039 (2018).
Yang, Y. G., Lindahl, T. & Barnes, D. E. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell 131, 873–886 (2007). [pii] 10.1016/j.cell.2007.10.017.
Stetson, D. B., Ko, J. S., Heidmann, T. & Medzhitov, R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 (2008).
Franz, K. M., Neidermyer, W. J., Tan, Y. J., Whelan, S. P. J. & Kagan, J. C. STING-dependent translation inhibition restricts RNA virus replication. Proc. Natl. Acad. Sci. USA 115, E2058–E2067 (2018).
Lindenbach, B. D. & Rice, C. M. Unravelling hepatitis C virus replication from genome to function. Nature 436, 933–938 (2005).
Meylan, E. et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172 (2005).
Nitta, S. et al. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology 57, 46–58 (2013).
Ding, Q. et al. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J. Hepatol. 59, 52–58 (2013).
Yi, G. et al. Hepatitis C virus NS4B can suppress STING accumulation to evade innate immune responses. J. Virol. 90, 254–265 (2016).
Rodenhuis-Zybert, I. A., Wilschut, J. & Smit, J. M. Dengue virus life cycle: viral and host factors modulating infectivity. Cell Mol. Life Sci. 67, 2773–2786 (2010).
Yu, C. Y. et al. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 8, e1002780 (2012).
Aguirre, S. et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 8, e1002934 (2012).
Aguirre, S. et al. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat. Microbiol 2, 17037 (2017).
Stabell, A.C. et al. Dengue viruses cleave STING in humans but not in nonhuman primates, their presumed natural reservoir. Elife 7, https://doi.org/10.7554/eLife.31919 (2018).
Bowen, J. R., Zimmerman, M. G. & Suthar, M. S. Taking the defensive: Immune control of Zika virus infection. Virus Res 254, 21–26 (2018).
Ding, Q. et al. Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc. Natl Acad. Sci. USA 115, E6310–E6318 (2018).
Zheng, Y. et al. Zika virus elicits inflammation to evade antiviral response by cleaving cGAS via NS1-caspase-1 axis. EMBO J 37, https://doi.org/10.15252/embj.201899347 (2018).
Liu, Y. & Cherry, S. Zika virus infection activates sting-dependent antiviral autophagy in the Drosophila brain. Autophagy 15, 174–175 (2019).
Coyne, C. B. STING’ing Zika virus in neurons. Nat. Microbiol 3, 975–976 (2018).
Goto, A. et al. The kinase IKKbeta regulates a STING- and NF-kappaB-dependent antiviral response pathway in drosophila. Immunity 49, 225–234 e224 (2018).
Tong, S. et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 9, e1003657 (2013).
Holm, C. K. et al. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat. Commun. 7, 10680 (2016).
Witte, C. E. et al. Innate immune pathways triggered by Listeria monocytogenes and their role in the induction of cell-mediated immunity. Adv. Immunol. 113, 135–156 (2012).
Archer, K. A., Durack, J. & Portnoy, D. A. STING-dependent type I IFN production inhibits cell-mediated immunity to Listeria monocytogenes. PLoS Pathog. 10, e1003861 (2014).
Koppe, U. et al. Streptococcus pneumoniae stimulates a STING- and IFN regulatory factor 3-dependent type I IFN production in macrophages, which regulates RANTES production in macrophages, cocultured alveolar epithelial cells, and mouse lungs. J. Immunol. 188, 811–817 (2012).
Parker, D. et al. Streptococcus pneumoniae DNA initiates type I interferon signaling in the respiratory tract. MBio 2, e00016–00011 (2011).
Mitzel, D. N., Lowry, V., Shirali, A. C., Liu, Y. & Stout-Delgado, H. W. Age-enhanced endoplasmic reticulum stress contributes to increased Atg9A inhibition of STING-mediated IFN-beta production during Streptococcus pneumoniae infection. J. Immunol. 192, 4273–4283 (2014).
Smith, I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol Rev. 16, 463–496 (2003).
Watson, R. O., Manzanillo, P. S. & Cox, J. S. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150, 803–815 (2012).
Manzanillo, P. S., Shiloh, M. U., Portnoy, D. A. & Cox, J. S. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11, 469–480 (2012).
Donovan, M. L., Schultz, T. E., Duke, T. J., Blumenthal, A. & Type, I. Interferons in the pathogenesis of tuberculosis: molecular drivers and immunological consequences. Front Immunol. 8, 1633 (2017).
Moreira-Teixeira, L., Mayer-Barber, K., Sher, A. & O’Garra, A. Type I interferons in tuberculosis: Foe and occasionally friend. J. Exp. Med. 215, 1273–1285 (2018).
Collins, A. C. et al. Cyclic GMP-AMP Synthase Is an Innate Immune DNA Sensor for Mycobacterium tuberculosis. Cell Host Microbe 17, 820–828 (2015).
Dey, B. et al. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat. Med 21, 401–406 (2015).
Marinho, F. V., Benmerzoug, S., Oliveira, S. C., Ryffel, B. & Quesniaux, V. F. J. The emerging roles of STING in bacterial infections. Trends Microbiol 25, 906–918 (2017).
Lindner, S. E., Miller, J. L. & Kappe, S. H. Malaria parasite pre-erythrocytic infection: preparation meets opportunity. Cell Microbiol 14, 316–324 (2012).
Miller, J. L., Sack, B. K., Baldwin, M., Vaughan, A. M. & Kappe, S. H. I. Interferon-mediated innate immune responses against malaria parasite liver stages. Cell Rep. 7, 436–447 (2014).
Sisquella, X. et al. Malaria parasite DNA-harbouring vesicles activate cytosolic immune sensors. Nat. Commun. 8, 1985 (2017).
Yu, X. et al. Cross-regulation of Two Type I interferon signaling pathways in plasmacytoid dendritic cells controls anti-malaria immunity and host mortality. Immunity 45, 1093–1107 (2016).
Wilson, D. R. et al. Biodegradable STING agonist nanoparticles for enhanced cancer immunotherapy. Nanomed.-Nanotechnol. 14, 237–246 (2018).
Corrales, L., Matson, V., Flood, B., Spranger, S. & Gajewski, T. F. Innate immune signaling and regulation in cancer immunotherapy. Cell Res 27, 96–108 (2017).
Fu, J. et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci. Transl. Med 7, 283ra252 (2015).
Ramanjulu, J. M. et al. Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature 564, 439–443 (2018).
Corrales, L. et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030 (2015).
Ahn, J., Xia, T., Rabasa Capote, A., Betancourt, D. & Barber, G. N. Extrinsic phagocyte-dependent STING signaling dictates the immunogenicity of dying cells. Cancer Cell 33, 862–873 e865 (2018).
Skouboe, M. K. et al. STING agonists enable antiviral cross-talk between human cells and confer protection against genital herpes in mice. PLoS Pathog. 14, e1006976 (2018).
Cavlar, T., Deimling, T., Ablasser, A., Hopfner, K. P. & Hornung, V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 32, 1440–1450 (2013).
Conlon, J. et al. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J. Immunol. 190, 5216–5225 (2013).
Guo, F. et al. STING agonists induce an innate antiviral immune response against hepatitis B virus. Antimicrob. Agents Chemother. 59, 1273–1281 (2015).
Gall, B. et al. Emerging alphaviruses are sensitive to cellular states induced by a novel small-molecule agonist of the STING pathway. J. Virol. 92, https://doi.org/10.1128/JVI.01913-17 (2018).
Sali, T. M. et al. Characterization of a novel human-specific STING agonist that elicits antiviral activity against emerging alphaviruses. PLoS Pathog. 11, e1005324 (2015).
Van Dis, E. et al. STING-activating adjuvants Elicit a Th17 immune response and protect against mycobacterium tuberculosis infection. Cell Rep. 23, 1435–1447 (2018).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ahn, J., Barber, G.N. STING signaling and host defense against microbial infection. Exp Mol Med 51, 1–10 (2019). https://doi.org/10.1038/s12276-019-0333-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-019-0333-0
This article is cited by
-
cGAS-STING, inflammasomes and pyroptosis: an overview of crosstalk mechanism of activation and regulation
Cell Communication and Signaling (2024)
-
STING dependent BAX-IRF3 signaling results in apoptosis during late-stage Coxiella burnetii infection
Cell Death & Disease (2024)
-
Attenuation of IFN signaling due to m6A modification of the host epitranscriptome promotes EBV lytic reactivation
Journal of Biomedical Science (2023)
-
Enhancing the immunogenicity of lipid-nanoparticle mRNA vaccines by adjuvanting the ionizable lipid and the mRNA
Nature Biomedical Engineering (2023)
-
Mycobacterium tuberculosis and its clever approaches to escape the deadly macrophage
World Journal of Microbiology and Biotechnology (2023)