Abstract
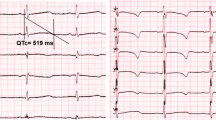
The recent introduction of genome sequencing in genetic analysis has led to the identification of pathogenic variants located in deep introns. Recently, several new tools have emerged to predict the impact of variants on splicing. Here, we present a Japanese boy of Joubert syndrome with biallelic TCTN2 variants. Exome sequencing identified only a heterozygous maternal nonsense TCTN2 variant (NM_024809.5:c.916C >T, p.(Gln306Ter)). Subsequent genome sequencing identified a deep intronic variant (c.1033+423G>A) inherited from his father. The machine learning algorithms SpliceAI, Squirls, and Pangolin were unable to predict alterations in splicing by the c.1033+423G>A variant. SpliceRover, a tool for splice site prediction using FASTA sequence, was able to detect a cryptic exon which was 85-bp away from the variant and within the inverted Alu sequence while SpliceRover scores for these splice sites showed slight increase (donor) or decrease (acceptor) between the reference and mutant sequences. RNA sequencing and RT-PCR using urinary cells confirmed inclusion of the cryptic exon. The patient showed major symptoms of TCTN2-related disorders such as developmental delay, dysmorphic facial features and polydactyly. He also showed uncommon features such as retinal dystrophy, exotropia, abnormal pattern of respiration, and periventricular heterotopia, confirming these as one of features of TCTN2-related disorders. Our study highlights usefulness of genome sequencing and RNA sequencing using urinary cells for molecular diagnosis of genetic disorders and suggests that database of cryptic splice sites predicted in introns by SpliceRover using the reference sequences can be helpful in extracting candidate variants from large numbers of intronic variants in genome sequencing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Lionel AC, Costain G, Monfared N, Walker S, Reuter MS, Hosseini SM, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med. 2018;20:435–43.
Vaz-Drago R, Custódio N, Carmo-Fonseca M. Deep intronic mutations and human disease. Hum Genet. 2017;136:1093–111.
Lee H, Huang AY, Wang LK, Yoon AJ, Renteria G, Eskin A, et al. Diagnostic utility of transcriptome sequencing for rare Mendelian diseases. Genet Med. 2020;22:490–9.
Hiraide T, Shimizu K, Miyamoto S, Aoto K, Nakashima M, Yamaguchi T, et al. Genome sequencing and RNA sequencing of urinary cells reveal an intronic FBN1 variant causing aberrant splicing. J Hum Genet. 2022;67:387–92.
Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535.e24.
Riepe TV, Khan M, Roosing S, Cremers FPM, Hoen PAC‘t. Benchmarking deep learning splice prediction tools using functional splice assays. Hum Mutat. 2021;42:799–810.
Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–74.
Zuallaert J, Godin F, Kim M, Soete A, Saeys Y, De Neve W. SpliceRover: interpretable convolutional neural networks for improved splice site prediction. Bioinformatics. 2018;34:4180–8.
Cheng J, Nguyen TYD, Cygan KJ, Çelik MH, Fairbrother WG, Avsec Ž, et al. MMSplice: modular modeling improves the predictions of genetic variant effects on splicing. Genome Biol. 2019;20:48.
Zeng T, Li YI. Predicting RNA splicing from DNA sequence using Pangolin. Genome Biol. 2022;23:103.
Danis D, Jacobsen JOB, Carmody LC, Gargano MA, McMurry JA, Hegde A, et al. Interpretable prioritization of splice variants in diagnostic next-generation sequencing. Am J Hum Genet. 2021;108:1564–77.
Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–84.
Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–28.
Huppke P, Wegener E, Böhrer-Rabel H, Bolz HJ, Zoll B, Gärtner J, et al. Tectonic gene mutations in patients with Joubert syndrome. Eur J Hum Genet. 2015;23:616–20.
Shaheen R, Faqeih E, Seidahmed MZ, Sunker A, Alali FE, AlQahtani K, et al. A TCTN2 mutation defines a novel Meckel Gruber syndrome locus. Hum Mutat. 2011;32:573–8.
Radha Rama Devi A, Naushad SM, Lingappa L. Clinical and molecular diagnosis of joubert syndrome and related disorders. Pediatr Neurol. 2020;106:43–9.
Van De Weghe JC, Gomez A, Doherty D. The Joubert-Meckel-nephronophthisis spectrum of ciliopathies. Annu Rev Genom Hum Genet. 2022;23:301–29.
Watanabe K, Nakashima M, Kumada S, Mashimo H, Enokizono M, Yamada K, et al. Identification of two novel de novo TUBB variants in cases with brain malformations: case reports and literature review. J Hum Genet. 2021;66:1193–7.
Tadaka S, Hishinuma E, Komaki S, Motoike IN, Kawashima J, Saigusa D, et al. jMorp updates in 2020: large enhancement of multi-omics data resources on the general Japanese population. Nucleic Acids Res. 2021;49:D536–44.
“GEM Japan Whole Genome Aggregation (GEM-J WGA) Panel”. Japan: GEnome Medical alliance Japan Project (GEM-J). Available from: https://togovar.biosciencedbc.jp/doc/datasets/gem_j_wga.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Wai HA, Lord J, Lyon M, Gunning A, Kelly H, Cibin P, et al. Blood RNA analysis can increase clinical diagnostic rate and resolve variants of uncertain significance. Genet Med. 2020;22:1005–14.
Strauch Y, Lord J, Niranjan M, Baralle D. CI-SpliceAI-Improving machine learning predictions of disease causing splicing variants using curated alternative splice sites. PLoS One. 2022;17:e0269159.
Jang W, Park J, Chae H, Kim M. Comparison of in silico tools for splice-altering variant prediction using established spliceogenic variants: an end-user’s point of view. Int J Genomics. 2022;2022:5265686.
Lee J, Jeong H, Won D, Shin S, Lee ST, Choi JR, et al. Noncanonical splice site and deep intronic FRMD7 variants activate cryptic exons in X-linked infantile nystagmus. Transl Vis Sci Technol. 2022;11:25.
Ram O, Schwartz S, Ast G. Multifactorial interplay controls the splicing profile of Alu-derived exons. Mol Cell Biol. 2008;28:3513–25.
Hiraide T, Nakashima M, Ikeda T, Tanaka D, Osaka H, Saitsu H. Identification of a deep intronic POLR3A variant causing inclusion of a pseudoexon derived from an Alu element in Pol III-related leukodystrophy. J Hum Genet. 2020;65:921–5.
Aicher JK, Jewell P, Vaquero-Garcia J, Barash Y, Bhoj EJ. Mapping RNA splicing variations in clinically accessible and nonaccessible tissues to facilitate Mendelian disease diagnosis using RNA-seq. Genet Med. 2020;22:1181–90.
Juric-Sekhar G, Adkins J, Doherty D, Hevner RF. Joubert syndrome: brain and spinal cord malformations in genotyped cases and implications for neurodevelopmental functions of primary cilia. Acta Neuropathol. 2012;123:695–709.
Bachmann-Gagescu R, Dempsey JC, Phelps IG, O’Roak BJ, Knutzen DM, Rue TC, et al. Joubert syndrome: a model for untangling recessive disorders with extreme genetic heterogeneity. J Med Genet. 2015;52:514–22.
Kars ME, Başak AN, Onat OE, Bilguvar K, Choi J, Itan Y, et al. The genetic structure of the Turkish population reveals high levels of variation and admixture. Proc Natl Acad Sci USA. 2021;118:e2026076118.
Al-Hamed MH, Kurdi W, Alsahan N, Alabdullah Z, Abudraz R, Tulbah M, et al. Genetic spectrum of Saudi Arabian patients with antenatal cystic kidney disease and ciliopathy phenotypes using a targeted renal gene panel. J Med Genet. 2016;53:338–47.
Shaheen R, Szymanska K, Basu B, Patel N, Ewida N, Faqeih E, et al. Characterizing the morbid genome of ciliopathies. Genome Biol. 2016;17:242.
Zhang M, Chang Z, Tian Y, Wang L, Lu Y. Two novel TCTN2 mutations cause Meckel-Gruber syndrome. J Hum Genet. 2020;65:1039–43.
Greenbaum L, Pode-Shakked B, Eisenberg-Barzilai S, Dicastro-Keidar M, Bar-Ziv A, Goldstein N, et al. Evaluation of diagnostic yield in fetal whole-exome sequencing: a report on 45 consecutive families. Front Genet. 2019;10:425.
Litz Philipsborn S, Hartmajer S, Shtorch Asor A, Vinovezky M, Regev M, Singer A, et al. A founder mutation in TCTN2 causes Meckel-Gruber syndrome type 8 among Jews of Ethiopian and Yemenite origin. Am J Med Genet A. 2021;185:1610–3.
Gana S, Serpieri V, Valente EM. Genotype-phenotype correlates in Joubert syndrome: a review. Am J Med Genet C Semin Med Genet. 2022;190:72–88.
Acknowledgements
We would like to thank the patients for participating in this work.
Funding
This work was supported by the Japan Agency for Medical Research and Development (AMED) (JP21ek0109549 to TO), Grants-in-Aid for Scientific Research (B) (JP20H03641), Grant-in-Aid for Challenging Research (Exploratory) (20K21570) from the Japan Society for the Promotion of Science, Japan Intractable Diseases (Nanbyo) Research Foundation (2020A02), and the Takeda Science Foundation.
Author information
Authors and Affiliations
Contributions
HS contributed to the conception and design of the study. TH, KS, SM, KA, MN, TY, TK, TO and HS contributed to the acquisition and analysis of data. TH, KS, and HS contributed to drafting the text and preparing the figure. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hiraide, T., Shimizu, K., Okumura, Y. et al. A deep intronic TCTN2 variant activating a cryptic exon predicted by SpliceRover in a patient with Joubert syndrome. J Hum Genet 68, 499–505 (2023). https://doi.org/10.1038/s10038-023-01143-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-023-01143-3
This article is cited by
-
RNA sequencing and target long-read sequencing reveal an intronic transposon insertion causing aberrant splicing
Journal of Human Genetics (2023)