Abstract
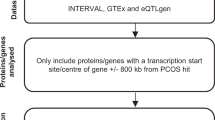
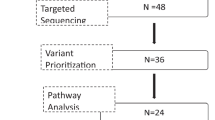
Polycystic ovary syndrome (PCOS) is a common endocrine disorder, which is accompanied by a variety of comorbidities including metabolic, reproductive, and psychiatric disorders. Genome-wide association studies have identified several genetic variants that are associated with PCOS. However, these variants often occur outside of coding regions and require further investigation to understand their contribution to PCOS. A transcriptome-wide association study (TWAS) was performed to uncover heritable gene expression profiles that are associated with PCOS in two independent cohorts. Causal gene prioritization was subsequently performed and expression of genes prioritized through these analyses was examined in 49 PCOS patients and 30 controls. TWAS analyses revealed that increased expression of ARL14EP was significantly associated with PCOS risk in the discovery (P = 1.6 × 10-6) and replication cohorts (P = 2.0 × 10-13). Gene prioritization pipelines provided further evidence that ARL14EP is the most likely causal gene at this locus. ARL14EP gene expression was shown to be significantly different between PCOS cases and controls, after adjusting for body mass index, age and testosterone levels (P = 1.2 × 10-13). This study has provided evidence for the role of ARL14EP in PCOS. Given that ARL14EP has been reported to play an important role in chromatin remodeling, variants affecting the expression of ARL14EP may also affect the expression of other genes that contribute to PCOS pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others

Data availability
Summary statistics utilized in this publication were obtained from https://www.repository.cam.ac.uk/handle/1810/283491.
Code availability
The code used in this manuscript is available at https://github.com/SarahMLyle/Identification-of-Gene-Expression-Patterns-Associated-with-Polycystic-Ovary-Syndrome-
References
Franks S. Polycystic ovary syndrome. N. Engl J Med. 1995;333:853–61.
Azziz R. Introduction: Determinants of polycystic ovary syndrome. Fertil Steril. 2016;106:4–5.
Deswal R, Narwal V, Dang A, Pundir CS. The prevalence of polycystic ovary syndrome: A brief systematic review. J Hum Reprod Sci. 2020;13:261–71.
Liu J, Wu Q, Hao Y, Jiao M, Wang X, Jiang S, et al. Measuring the global disease burden of polycystic ovary syndrome in 194 countries: Global Burden of Disease Study 2017. Hum Reprod. 2021;36:1108–19.
Huddleston HG, Dokras A. Diagnosis and treatment of polycystic ovary syndrome. JAMA - J Am Med Assoc. 2022;327:274–5.
Williams T, Mortada R, Porter S. Diagnosis and treatment of polycystic ovary syndrome. Am Fam Physician. 2016;94:106–13.
Vink JM, Sadrzadeh S, Lambalk CB, Boomsma DI. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab. 2006;91:2100–4.
Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Shi Y, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet 2011;43:55–9.
Shi Y, Zhao H, Shi Y, Cao Y, Yang D, Li Z, et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. 2012;44:1020–5.
Hayes MG, Urbanek M, Ehrmann DA, Armstrong LL, Lee JY, Sisk R, et al. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nature. Communications 2015;6:14.
Day FR, Hinds DA, Tung JY, Stolk L, Styrkarsdottir U, Saxena R, et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat Commun. 2015;6:1–7.
Day F, Karaderi T, Jones MR, Meun C, He C, Drong A, et al. Large-scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PLoS Genet. 2018;14:1–20.
Barbeira AN, Bonazzola R, Gamazon ER, Liang Y, Park YS, Kim-Hellmuth S, et al. Exploiting the GTEx resources to decipher the mechanisms at GWAS loci. Genome Biol. 2021;22:1–24.
Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D, et al. Opportunities and challenges for transcriptome-wide association studies. Nat Genet. 2019;51:592–9.
Pividori M, Rajagopal PS, Barbeira A, Liang Y, Melia O, Bastarache L, et al. PhenomeXcan: Mapping the genome to the phenome through the transcriptome. Sci Adv. 2020;6:eaba2083.
Doke T, Huang S, Qiu C, Liu H, Guan Y, Hu H, et al. Transcriptome-wide association analysis identifies DACH1 as a kidney disease risk gene that contributes to fibrosis. J Clin Investig. 2021;1:131.
Wu C, Tan S, Liu L, Cheng S, Li P, Li W, et al. Transcriptome-wide association study identifies susceptibility genes for rheumatoid arthritis. Arthr Res Ther. 2021;1:23.
Li X, Su X, Liu J, Li H, Li M, Li W, et al. Transcriptome-wide association study identifies new susceptibility genes and pathways for depression. Transl Psychiatry. 2021;1:11.
Pan C, Ning Y, Jia Y, Cheng S, Wen Y, Yang X, et al. Transcriptome-wide association study identified candidate genes associated with gut microbiota. Gut Pathog. 2021;1:13.
Hormozdiari F, Kichaev G, Yang WY, Pasaniuc B, Eskin E. Identification of causal genes for complex traits. Bioinformatics 2015;31:i206–13.
Mancuso N, Freund MK, Johnson R, Shi H, Kichaev G, Gusev A, et al. Probabilistic fine-mapping of transcriptome-wide association studies. Nat Genet. 2019;51:675–82.
Gazal S, Weissbrod O, Hormozdiari F, Dey KK, Nasser J, Jagadeesh KA, et al. Combining SNP-to-gene linking strategies to identify disease genes and assess disease omnigenicity. Nat Genet. 2022;54:827–36.
Romm A. PCOS Summary Sheet [Internet]. [cited 2022 Jul 7]. Available from: https://www.repository.cam.ac.uk/handle/1810/283491
GWAS. Harmonization And Imputation· hakyimlab/summary-gwas-imputation Wiki [Internet]. GitHub. [cited 2022 Nov 20]. Available from: https://github.com/hakyimlab/summary-gwas-imputation
Auton A, Abecasis GR, Altshuler DM, Durbin RM, Bentley DR, Chakravarti A, et al. A global reference for human genetic variation. Vol. 526, Nature. Nature Publishing Group; 2015. p. 68–74.
GTEx Project. GTEx portal. GTEx Analysis Release V6p (dbGaP Accession phs000424.v6.p1). 2017.
Barbeira AN, Pividori MD, Zheng J, Wheeler HE, Nicolae DL, Im HK. Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet. 2019;15:e1007889.
FinnGen results [Internet]. [cited 2022 Nov 20]. Available from: https://r7.finngen.fi/pheno/E4_PCOS_CONCORTIUM
Liao C, Laporte AD, Spiegelman D, Akçimen F, Joober R, Dion PA, et al. Transcriptome-wide association study of attention deficit hyperactivity disorder identifies associated genes and phenotypes.
Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLOS Genet. 2014;10:e1004383.
Rentzsch P, Schubach M, Shendure J, Kircher M. CADD-Splice—improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;1:13.
Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, et al. Ensembl 2022. Nucleic Acids Res. 2022;50:D988–95.
Watanabe K, Taskesen E, Van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nature. Communications 2017;8:1–11.
LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants - PMC [Internet]. [cited 2022 Dec 9]. Available from: https://www-ncbi-nlm-nih-gov.uml.idm.oclc.org/pmc/articles/PMC4626747/
Kokosar M, Benrick A, Perfilyev A, Fornes R, Nilsson E, Maliqueo M, et al. Epigenetic and transcriptional alterations in human adipose tissue of polycystic ovary syndrome. Sci Rep. 2016;6:22883.
Stener-Victorin E, Holm G, Labrie F, Nilsson L, Janson PO, Ohlsson C. Are there any sensitive and specific sex steroid markers for polycystic ovary syndrome? J Clin Endocrinol Metab. 2010;95:810–9.
Alkes Group [Internet]. [cited 2022 Nov 20]. Available from: https://alkesgroup.broadinstitute.org/cS2G/code
Misgar RA, Wani AI, Bankura B, Bashir MI, Roy A, Das M. FSH β-subunit mutations in two sisters: the first report from the Indian sub-continent and review of previous cases. Gynecol Endocrinol. 2019;35:290–3.
Li Y, Chen C, Ma Y, Xiao J, Luo G, Li Y, et al. Multi-system reproductive metabolic disorder: significance for the pathogenesis and therapy of polycystic ovary syndrome (PCOS). Vol. 228, Life Sciences. Elsevier Inc.; 2019. p. 167–75.
Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118.
OMIM Entry - * 612295 - ADP-RIBOSYLATION FACTOR-LIKE GTPase 14 EFFECTOR PROTEIN; ARL14EP [Internet]. [cited 2022 May 12]. Available from: https://omim.org/entry/612295
OMIM Entry - # 194072 - WILMS TUMOR, ANIRIDIA, GENITOURINARY ANOMALIES, AND MENTAL RETARDATION SYNDROME; WAGR [Internet]. [cited 2022 May 12]. Available from: https://omim.org/entry/194072
Duffy KA, Trout KL, Gunckle JM, Krantz SMC, Morris J, Kalish JM. Results From the WAGR Syndrome Patient Registry: Characterization of WAGR Spectrum and Recommendations for Care Management. Front Pediatrics. 2021;9:733018.
Peter CJ, Saito A, Hasegawa Y, Tanaka Y, Nagpal M, Perez G, et al. In vivo epigenetic editing of Sema6a promoter reverses transcallosal dysconnectivity caused by C11orf46/Arl14ep risk gene. Nat Commun. 2019;10:4112.
Sakaue S, Weinand K, Dey KK, Jagadeesh K, Kanai M, Watts GFM, et al. Tissue-specific enhancer-gene maps from multimodal single-cell data identify causal disease alleles [Internet]. Genet Genom Med. 2022 [cited 2023 Jan 3]. Available from: http://medrxiv.org/lookup/doi/10.1101/2022.10.27.22281574
Zhang Z, Zamojski M, Smith GR, Willis TL, Yianni V, Mendelev N, et al. Single nucleus transcriptome and chromatin accessibility of postmortem human pituitaries reveal diverse stem cell regulatory mechanisms. Cell Rep. 2022;38:110467.
Censin JC, Bovijn J, Holmes M, Lindgren C. Colocalization analysis of polycystic ovary syndrome to identify potential disease-mediating genes and proteins. Eur J Human Genet. 2021;29:1446–54.
Sen A, Prizant H, Light A, Biswas A, Hayes E, Lee HJ, et al. Androgens regulate ovarian follicular development by increasing follicle stimulating hormone receptor and microRNA-125b expression. Proc Natl Acad Sci USA. 2014;111:3008–13.
Acknowledgements
Galen EB Wright provided critical feedback during the writing process.
Funding
BID is supported by a CIHR Tier 2 Canada Research Chair in Pharmacogenomics. SML was supported by BioTalent Canada. SA was supported by Research Manitoba and NSERC VADA.
Author information
Authors and Affiliations
Contributions
SML: Performed the analysis; Wrote the paper. SA: Performed analyses. JE: Wrote the paper. ES-V: Provided data. MN: Wrote the paper. BID: Conceived and designed the analysis; Wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Ethical approvals were not required for the TWAS, as theses analyses were performed using publicly available summary statistics. The summary statistics obtained did not include identifiable participant information and thus ensured the privacy of participants. Ethical approval for the targeted gene expression analyses was obtained from the Regional Ethical Review Board of the University of Gothenburg in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lyle, S.M., Ahmed, S., Elliott, J.E. et al. Transcriptome-wide association analyses identify an association between ARL14EP and polycystic ovary syndrome. J Hum Genet 68, 347–353 (2023). https://doi.org/10.1038/s10038-023-01120-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-023-01120-w