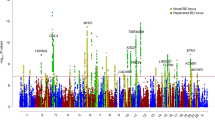
Abstract
Bipolar disorder (BD) is a common mental disorder characterized by recurrent mood episodes, which causes major socioeconomic burdens globally. Though its disease pathogenesis is largely unknown, the high heritability of BD indicates strong contributions from genetic factors. In this review, we summarize the recent achievements in the genetics of BD, particularly those from genome-wide association study (GWAS) of common variants and next-generation sequencing analysis of rare variants. These include the identification of dozens of robust disease-associated loci, deepening of our understanding of the biology of BD, objective description of correlations with other psychiatric disorders and behavioral traits, formulation of methods for predicting disease risk and drug response, and the discovery of a single gene associated with bipolar disorder and schizophrenia spectrum with a large effect size. On the other hand, the findings to date have not yet made a clear contribution to the improvement of clinical psychiatry of BD. We overview the remaining challenges as well as possible paths to resolve them, referring to studies of other major neuropsychiatric disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Grande I, Berk M, Birmaher B, Vieta E. Bipolar disorder. Lancet. 2016;387:1561–72.
Angst J, Sellaro R. Historical perspectives and natural history of bipolar disorder. Biol Psychiatry. 2000;48:445–57.
Michalak EE, Yatham LN, Lam RW. Quality of life in bipolar disorder: A review of the literature. Health Qual Life Outcomes. 2005;3:72.
Collaborators GBDMD. Global, regional, and national burden of 12 mental disorders in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Psychiatry. 2022;9:137–50.
Perlis RH, Miyahara S, Marangell LB, Wisniewski SR, Ostacher M, DelBello MP, et al. Long-Term implications of early onset in bipolar disorder: data from the first 1000 participants in the systematic treatment enhancement program for bipolar disorder (STEP-BD). Biol Psychiatry. 2004;55:875–81.
Lin P-I, McInnis MG, Potash JB, Willour V, MacKinnon DF, DePaulo JR, et al. Clinical correlates and familial aggregation of age at onset in bipolar disorder. Am J Psychiatry. 2006;163:240–6.
Nishioka M, Kazuno A-A, Nakamura T, Sakai N, Hayama T, Fujii K, et al. Systematic analysis of exonic germline and postzygotic de novo mutations in bipolar disorder. Nat Commun. 2021;12:3750.
Administration SAMHS. CBHSQ Methodology Report. In: Impact of the DSM-IV to DSM-5 Changes on the National Survey on Drug Use and Health. Rockville (MD): Substance Abuse and Mental Health Services Administration (US); 2016.
Diagnostic and statistical manual of mental disorders: DSM-5™. 5th edition. ed. American Psychiatric Publishing, a division of American Psychiatric Association: Washington, DC, 2013.
Clemente AS, Diniz BS, Nicolato R, Kapczinski FP, Soares JC, Firmo JO, et al. Bipolar disorder prevalence: a systematic review and meta-analysis of the literature. Rev Brasileira de Psiquiatria (Sao Paulo, Braz: 1999). 2015;37:155–61.
Craddock N, Jones I. Genetics of bipolar disorder. J Med Genet. 1999;36:585–94.
Merikangas KR, Jin R, He JP, Kessler RC, Lee S, Sampson NA, et al. Prevalence and correlates of bipolar spectrum disorder in the world mental health survey initiative. Arch Gen Psychiatry. 2011;68:241–51.
Kato T, Baba K, Guo W, Chen Y, Nosaka T. Impact of bipolar disorder on health-related quality of life and work productivity: Estimates from the national health and wellness survey in Japan. J Affect Disord. 2021;295:203–14.
Nishi D, Ishikawa H, Kawakami N. Prevalence of mental disorders and mental health service use in Japan. Psychiatry Clin Neurosci. 2019;73:458–65. https://doi.org/10.1111/pcn.12894.
Kieseppä T, Partonen T, Haukka J, Kaprio J, Lönnqvist J. High concordance of bipolar I disorder in a nationwide sample of twins. Am J Psychiatry. 2004;161:1814–21.
Khalilisamani N, Thomson PC, Raadsma HW, Khatkar MS. Estimating heritability using family-pooled phenotypic and genotypic data: a simulation study applied to aquaculture. Heredity. 2022;128:178–86.
Barnett JH, Smoller JW. The genetics of bipolar disorder. Neuroscience. 2009;164:331–43.
Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet Part C, Semin Med Genet. 2003;123c:48–58.
Wender PH, Kety SS, Rosenthal D, Schulsinger F, Ortmann J, Lunde I. Psychiatric disorders in the biological and adoptive families of adopted individuals with affective disorders. Arch Gen Psychiatry. 1986;43:923–29.
Parboosing R, Bao Y, Shen L, Schaefer CA, Brown AS. Gestational influenza and bipolar disorder in adult offspring. JAMA Psychiatry. 2013;70:677–85.
Talati A, Bao Y, Kaufman J, Shen L, Schaefer CA, Brown AS. Maternal smoking during pregnancy and bipolar disorder in offspring. Am J Psychiatry. 2013;170:1178–85.
Aas M, Henry C, Andreassen OA, Bellivier F, Melle I, Etain B. The role of childhood trauma in bipolar disorders. Int J Bipolar Disord. 2016;4:2–2.
Pulst SM. Genetic linkage analysis. Arch Neurol. 1999;56:667–72.
Jones I, Craddock N. Candidate gene studies of bipolar disorder. Ann Med. 2001;33:248–56.
Heiden A, Schüssler P, Itzlinger U, Leisch F, Scharfetter J, Gebhardt C, et al. Association studies of candidate genes in bipolar disorders. Neuropsychobiology. 2000;42:18–21.
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J. et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921.
Consortium IH. The international HapMap project. Nature. 2003;426:789–96.
Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–5.
Psychiatric GCBDWG. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–83.
Baum A, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207.
Charney A, Ruderfer D, Stahl E, Moran J, Chambert K, Belliveau R, et al. Evidence for genetic heterogeneity between clinical subtypes of bipolar disorder. Transl Psychiatry. 2017;7:e993–e93.
Chen D, Jiang X, Akula N, Shugart Y, Wendland J, Steele C, et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry. 2013;18:195–205.
Ikeda M, Takahashi A, Kamatani Y, Okahisa Y, Kunugi H, Mori N, et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol Psychiatry. 2018;23:639–47.
Hou L, Bergen SE, Akula N, Song J, Hultman CM, Landén M, et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet. 2016;25:3383–94.
Mühleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun. 2014;5:1–8.
Green EK, Grozeva D, Forty L, Gordon-Smith K, Russell E, Farmer A, et al. Association at SYNE1 in both bipolar disorder and recurrent major depression. Mol Psychiatry. 2013;18:614–17.
Green EK, Hamshere M, Forty L, Gordon-Smith K, Fraser C, Russell E, et al. Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case–control sample. Mol Psychiatry. 2013;18:1302–07.
Cichon S, Mühleisen TW, Degenhardt FA, Mattheisen M, Miró X, Strohmaier J, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88:372–81.
Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–8.
Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci. 2009;106:7501–06.
Sklar P, Smoller JW, Fan J, Ferreira MAR, Perlis RH, Chambert K, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13:558–69.
Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78.
Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry. 2009;14:755–63.
Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803.
Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–29.
Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–08.
Lam M, Chen C-Y, Li Z, Martin AR, Bryois J, Ma X, et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet. 2019;51:1670–78.
Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7.
Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–59.
Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–76.
Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52.
Jia X, Goes FS, Locke AE, Palmer D, Wang W, Cohen-Woods S, et al. Investigating rare pathogenic/likely pathogenic exonic variation in bipolar disorder. Mol Psychiatry. 2021;26:5239–50.
Palmer DS, Howrigan DP, Chapman SB, Adolfsson R, Bass N, Blackwood D, et al. Exome sequencing in bipolar disorder identifies AKAP11 as a risk gene shared with schizophrenia. Nat Genet. 2022;54:541–7.
Goes FS, Pirooznia M, Parla JS, Kramer M, Ghiban E, Mavruk S, et al. Exome sequencing of familial bipolar disorder. JAMA Psychiatry. 2016;73:590–97.
Husson T, Duboc J-B, Quenez O, Charbonnier C, Rotharmel M, Cuenca M, et al. Identification of potential genetic risk factors for bipolar disorder by whole-exome sequencing. Transl Psychiatry. 2018;8:268.
Kataoka M, Matoba N, Sawada T, Kazuno AA, Ishiwata M, Fujii K, et al. Exome sequencing for bipolar disorder points to roles of de novo loss-of-function and protein-altering mutations. Mol Psychiatry. 2016;21:885–93.
Lescai F, Als TD, Li Q, Nyegaard M, Andorsdottir G, Biskopstø M, et al. Whole-exome sequencing of individuals from an isolated population implicates rare risk variants in bipolar disorder. Transl Psychiatry. 2017;7:e1034–e34.
Ament Seth A, Szelinger S, Glusman G, Ashworth J, Hou L, Akula N, et al. Rare variants in neuronal excitability genes influence risk for bipolar disorder. Proc Natl Acad Sci. 2015;112:3576–81.
Charney AW, Stahl EA, Green EK, Chen C-Y, Moran JL, Chambert K, et al. Contribution of rare copy number variants to bipolar disorder risk is limited to schizoaffective cases. Biol Psychiatry. 2019;86:110–19.
Green EK, Rees E, Walters JTR, Smith KG, Forty L, Grozeva D, et al. Copy number variation in bipolar disorder. Mol Psychiatry. 2016;21:89–93.
Grozeva D, Kirov G, Ivanov D, Jones IR, Jones L, Green EK, et al. Rare copy number variants: a point of rarity in genetic risk for bipolar disorder and schizophrenia. Arch Gen Psychiatry. 2010;67:318–27.
Calle Sánchez X, Helenius D, Bybjerg-Grauholm J, Pedersen C, Hougaard DM, Børglum AD, et al. Comparing copy number variations in a danish case cohort of individuals with psychiatric disorders. JAMA Psychiatry. 2022;79:59–69.
Zhang D, Cheng L, Qian Y, Alliey-Rodriguez N, Kelsoe JR, Greenwood T, et al. Singleton deletions throughout the genome increase risk of bipolar disorder. Mol Psychiatry. 2009;14:376–80.
Noor A, Lionel AC, Cohen-Woods S, Moghimi N, Rucker J, Fennell A, et al. Copy number variant study of bipolar disorder in Canadian and UK populations implicates synaptic genes. Am J Med Genet Part B: Neuropsychiatr Genet. 2014;165:303–13. https://doi.org/10.1002/ajmg.b.32232.
Priebe L, Degenhardt F, Strohmaier J, Breuer R, Herms S, Witt SH, et al. Copy number variants in german patients with schizophrenia. PLOS ONE. 2013;8:e64035.
Bergen SE, O’Dushlaine CT, Ripke S, Lee PH, Ruderfer DM, Akterin S, et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry. 2012;17:880–6.
Malhotra D, McCarthy S, Michaelson JJ, Vacic V, Burdick KE, Yoon S, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72:951–63.
Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLOS Computational Biol. 2015;11:e1004219.
Brainstorm C, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757.
Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–7.
Bipolar D.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Electronic address drve, Bipolar D, Schizophrenia Working Group of the Psychiatric Genomics C. Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell. 2018;173:1705–15.e16.
Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Communications 2018;9:224.
Kishi T, Ikuta T, Matsuda Y, Sakuma K, Okuya M, Mishima K, et al. Mood stabilizers and/or antipsychotics for bipolar disorder in the maintenance phase: a systematic review and network meta-analysis of randomized controlled trials. Mol Psychiatry. 2021;26:4146–57.
Yildiz A, Vieta E, Leucht S, Baldessarini RJ. Efficacy of antimanic treatments: meta-analysis of randomized, controlled trials. Neuropsychopharmacology. 2011;36:375–89.
Thase ME, Macfadden W, Weisler RH, Chang W, Paulsson B, Khan A, et al. Efficacy of quetiapine monotherapy in bipolar I and II depression: a double-blind, placebo-controlled study (the BOLDER II study). J Clin Psychopharmacol. 2006;26:600–9.
Young AH, McElroy SL, Olausson B, Paulsson B. A randomised, placebo-controlled 52-week trial of continued quetiapine treatment in recently depressed patients with bipolar I and bipolar II disorder. World J Biol Psychiatry. 2014;15:96–112.
Tohen M, McDonnell DP, Case M, Kanba S, Ha K, Fang YR, et al. Randomised, double-blind, placebo-controlled study of olanzapine in patients with bipolar I depression. Br J Psychiatry. 2012;201:376–82.
Tohen M, Calabrese JR, Sachs GS, Banov MD, Detke HC, Risser R, et al. Randomized, placebo-controlled trial of olanzapine as maintenance therapy in patients with bipolar I disorder responding to acute treatment with olanzapine. Am J Psychiatry. 2006;163:247–56.
Vieta E, Montgomery S, Sulaiman AH, Cordoba R, Huberlant B, Martinez L, et al. A randomized, double-blind, placebo-controlled trial to assess prevention of mood episodes with risperidone long-acting injectable in patients with bipolar I disorder. Eur Neuropsychopharmacol. 2012;22:825–35.
National Institute of Health and Care Excellence N. Bipolar disorder: assessment and management (2014, updated 2020), 2020.
Garnham J, Munro A, Slaney C, MacDougall M, Passmore M, Duffy A, et al. Prophylactic treatment response in bipolar disorder: Results of a naturalistic observation study. J Affect Disord. 2007;104:185–90.
Hou L, Heilbronner U, Degenhardt F, Adli M, Akiyama K, Akula N, et al. Genetic variants associated with response to lithium treatment in bipolar disorder: a genome-wide association study. Lancet. 2016;387:1085–93.
International Consortium on Lithium G. Association of polygenic score for schizophrenia and HLA antigen and inflammation genes with response to lithium in bipolar affective disorder: a genome-wide association study. JAMA Psychiatry. 2018;75:65–74.
Chung W-H, Hung S-I, Hong H-S, Hsih M-S, Yang L-C, Ho H-C, et al. A marker for Stevens–Johnson syndrome. Nature. 2004;428:486–86.
Tangamornsuksan W, Chaiyakunapruk N, Somkrua R, Lohitnavy M, Tassaneeyakul W. Relationship between the HLA-B*1502 allele and carbamazepine-induced stevens-johnson syndrome and toxic epidermal necrolysis: a systematic review and meta-analysis. JAMA Dermatol. 2013;149:1025–32.
Dean L. Carbamazepine Therapy and HLA Genotype. In: Pratt VM, Scott SA, Pirmohamed M, Esquivel B, Kane MS, Kattman BL, et al. eds. Medical Genetics Summaries. Bethesda (MD): National Center for Biotechnology Information (US); 2012.
Nicoletti P, Barrett S, McEvoy L, Daly AK, Aithal G, Lucena MI, et al. Shared genetic risk factors across carbamazepine-induced hypersensitivity reactions. Clin Pharmacol Therapeutics. 2019;106:1028–36.
Jang HW, Kim SW, Cho Y-J, Heo K, Lee BI, Lee SK, et al. GWAS identifies two susceptibility loci for lamotrigine-induced skin rash in patients with epilepsy. Epilepsy Res. 2015;115:88–94.
Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AAE, Lee SH, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47:1114–20.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Liu D, Meyer D, Fennessy B, Feng C, Cheng E, Johnson JS, et al. Rare schizophrenia risk variant burden is conserved in diverse human populations. medRxiv. 2022:2022.01.03.22268662.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23.
Li Q, Wang K. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100:267–80.
Deciphering Developmental Disorders S. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–8.
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21.
Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582–8.
Koemans TS, Kleefstra T, Chubak MC, Stone MH, Reijnders MRF, de Munnik S, et al. Functional convergence of histone methyltransferases EHMT1 and KMT2C involved in intellectual disability and autism spectrum disorder. PLOS Genet. 2017;13:e1006864.
Hood Rebecca L, Lines Matthew A, Nikkel Sarah M, Schwartzentruber J, Beaulieu C, Nowaczyk Małgorzata JM, et al. Mutations in SRCAP, encoding SNF2-related CREBBP activator protein, cause floating-harbor syndrome. Am J Hum Genet. 2012;90:308–13.
Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Sci (N. Y, NY). 2007;316:445–9.
Loh P-R, Genovese G, Handsaker RE, Finucane HK, Reshef YA, Palamara PF, et al. Insights into clonal haematopoiesis from 8342 mosaic chromosomal alterations. Nature. 2018;559:350–5.
Singh T, Poterba T, Curtis D, Akil H, Al Eissa M, Barchas JD, et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature. 2022;604:509–16.
Fu JM, Satterstrom FK, Peng M, Brand H, Collins RL, Dong S, et al. Rare coding variation illuminates the allelic architecture, risk genes, cellular expression patterns, and phenotypic context of autism. medRxiv. 2021:2021.12.20.21267194.
Takata A. Estimating contribution of rare non-coding variants to neuropsychiatric disorders. Psychiatry Clin Neurosci. 2019;73:2–10. https://doi.org/10.1111/pcn.12774.
Takata A, Ionita-Laza I, Gogos Joseph A, Xu B, Karayiorgou M. De novo synonymous mutations in regulatory elements contribute to the genetic etiology of autism and schizophrenia. Neuron. 2016;89:940–47.
Krupp DR, Barnard RA, Duffourd Y, Evans SA, Mulqueen RM, Bernier R, et al. Exonic mosaic mutations contribute risk for autism spectrum disorder. Am J Hum Genet. 2017;101:369–90.
Short PJ, McRae JF, Gallone G, Sifrim A, Won H, Geschwind DH, et al. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature. 2018;555:611–16.
Halvorsen M, Huh R, Oskolkov N, Wen J, Netotea S, Giusti-Rodriguez P, et al. Increased burden of ultra-rare structural variants localizing to boundaries of topologically associated domains in schizophrenia. Nat Commun. 2020;11:1842.
Alexeyev M, Shokolenko I, Wilson G, LeDoux S. The maintenance of mitochondrial DNA integrity–critical analysis and update. Cold Spring Harb Perspect Biol. 2013;5:a012641–a41.
Mitra I, Huang B, Mousavi N, Ma N, Lamkin M, Yanicky R. et al. Patterns of de novo tandem repeat mutations and their role in autism. Nature. 2021;589:246–50.
Trost B, Engchuan W, Nguyen CM, Thiruvahindrapuram B, Dolzhenko E, Backstrom I, et al. Genome-wide detection of tandem DNA repeats that are expanded in autism. Nature. 2020;586:80–6.
Mukamel RE, Handsaker RE, Sherman MA, Barton AR, Zheng Y, McCarroll SA, et al. Protein-coding repeat polymorphisms strongly shape diverse human phenotypes. Science. 2021;373:1499–505.
Schaefer GB, Mendelsohn NJ, for the Professional P, Guidelines C. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med. 2013;15:399–407.
Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism. 2013;4:36–36.
Singh T, Neale BM, Daly MJ, on behalf of the Schizophrenia Exome Meta-Analysis C. Exome sequencing identifies rare coding variants in 10 genes which confer substantial risk for schizophrenia. medRxiv. 2020:2020.09.18.20192815.
Achilly NP, Wang W, Zoghbi HY. Presymptomatic training mitigates functional deficits in a mouse model of Rett syndrome. Nature. 2021;592:596–600.
Mukai J, Cannavò E, Crabtree GW, Sun Z, Diamantopoulou A, Thakur P, et al. Recapitulation and reversal of schizophrenia-related phenotypes in Setd1a-deficient mice. Neuron. 2019;104:471–87.e12.
Chen Y, Yu J, Niu Y, Qin D, Liu H, Li G. et al. Modeling rett syndrome using TALEN-edited MECP2 mutant cynomolgus monkeys. Cell. 2017;169:945–55.e10.
Zhou Y, Sharma J, Ke Q, Landman R, Yuan J, Chen H. et al. Atypical behaviour and connectivity in SHANK3-mutant macaques. Nature. 2019;570:326–31.
Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, et al. FOXG1-dependent dysregulation of GABA/Glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–90.
Paulsen B, Velasco S, Kedaigle AJ, Pigoni M, Quadrato G, Deo AJ, et al. Autism genes converge on asynchronous development of shared neuron classes. Nature. 2022;602:268–73.
Elliott J, Bodinier B, Bond TA, Chadeau-Hyam M, Evangelou E, Moons KGM, et al. Predictive accuracy of a polygenic risk score–enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA. 2020;323:636–45.
Natarajan P, Young R, Stitziel NO, Padmanabhan S, Baber U, Mehran R, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135:2091–101.
Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–24.
Landi I, Kaji DA, Cotter L, Van Vleck T, Belbin G, Preuss M, et al. Prognostic value of polygenic risk scores for adults with psychosis. Nat Med. 2021;27:1576–81.
Liebers DT, Pirooznia M, Ganna A, Goes FS. Discriminating bipolar depression from major depressive disorder with polygenic risk scores. Psychol Med. 2021;51:1451–8.
Fiedorowicz JG, Endicott J, Leon AC, Solomon DA, Keller MB, Coryell WH. Subthreshold hypomanic symptoms in progression from unipolar major depression to bipolar disorder. Am J Psychiatry. 2011;168:40–8.
Funding
This work was in part supported by grants from the Japan Agency for Medical Research and Development (AMED) under Grant Number JP21km0405214 (to A.T.) and JSPS KAKENHI under Grant Numbers JP20H05777 (to A.T.), 21H02855 (to A.T.) and 20KK0225 (to Y.O). This study was also supported by the RIKEN Junior Research Associate Program (to T.H.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hara, T., Owada, Y. & Takata, A. Genetics of bipolar disorder: insights into its complex architecture and biology from common and rare variants. J Hum Genet 68, 183–191 (2023). https://doi.org/10.1038/s10038-022-01046-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-022-01046-9