Abstract
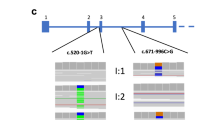
Exome sequencing and panel testing have improved diagnostic yield in genetic analysis by comprehensively detecting pathogenic variants in exonic regions. However, it is important to identify non-exonic pathogenic variants to further improve diagnostic yield. Here, we present a female proband and her father who is diagnosed with Marfan syndrome, a systemic connective tissue disorder caused by pathogenic variants in FBN1. There are also two affected individuals in the siblings of the father, indicating the genetic basis in this family. However, panel testing performed by two institutions reported no causal variants. To further explore the genetic basis of the family, we performed genome sequencing of the proband and RNA sequencing of urinary cells derived from urine samples of the proband and her father because FBN1 is strongly expressed in urinary cells though it is poorly expressed in peripheral blood mononuclear cells. Genome sequencing identified a rare intronic variant (c.5789-15G>A) in intron 47 of FBN1 (NM_000138.4), which was transmitted from her father. RNA sequencing revealed allelic imbalance (monoallelic expression) of FBN1, retention of intron 47, and fewer aberrant transcripts utilizing new acceptor sites within exon 48, which were confirmed by RT-PCR. These results highlighted urinary cells as clinically accessible tissues for RNA sequencing if disease-causing genes are not sufficiently expressed in the blood, and the usefulness of multi-omics analysis for molecular diagnosis of genetic disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sakai LY, Keene DR, Renard M, De Backer J. FBN1: The disease-causing gene for Marfan syndrome and other genetic disorders. Gene 2016;591:279–91.
Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–9.
Schwarze K, Buchanan J, Taylor JC, Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018;20:1122–30.
Lionel AC, Costain G, Monfared N, Walker S, Reuter MS, Hosseini SM, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med. 2018;20:435–43.
Maddirevula S, Kuwahara H, Ewida N, Shamseldin HE, Patel N, Alzahrani F, et al. Analysis of transcript-deleterious variants in Mendelian disorders: implications for RNA-based diagnostics. Genome Biol. 2020;21:145.
Kremer LS, Bader DM, Mertes C, Kopajtich R, Pichler G, Iuso A, et al. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat Commun. 2017;8:15824.
Cummings BB, Marshall JL, Tukiainen T, Lek M, Donkervoort S, Foley AR, et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci Transl Med. 2017;9:eaal5209.
Hamanaka K, Miyatake S, Koshimizu E, Tsurusaki Y, Mitsuhashi S, Iwama K, et al. RNA sequencing solved the most common but unrecognized NEB pathogenic variant in Japanese nemaline myopathy. Genet Med. 2019;21:1629–38.
Frésard L, Smail C, Ferraro NM, Teran NA, Li X, Smith KS, et al. Identification of rare-disease genes using blood transcriptome sequencing and large control cohorts. Nat Med. 2019;25:911–9.
Gonorazky HD, Naumenko S, Ramani AK, Nelakuditi V, Mashouri P, Wang P, et al. Expanding the boundaries of RNA sequencing as a diagnostic tool for rare mendelian disease. Am J Hum Genet. 2019;104:466–83.
Aicher JK, Jewell P, Vaquero-Garcia J, Barash Y, Bhoj EJ. Mapping RNA splicing variations in clinically accessible and nonaccessible tissues to facilitate Mendelian disease diagnosis using RNA-seq. Genet Med. 2020;22:1181–90.
Bronstein R, Capowski EE, Mehrotra S, Jansen AD, Navarro-Gomez D, Maher M, et al. A combined RNA-seq and whole genome sequencing approach for identification of non-coding pathogenic variants in single families. Hum Mol Genet. 2020;29:967–79.
Murdock DR, Dai H, Burrage LC, Rosenfeld JA, Ketkar S, Müller MF, et al. Transcriptome-directed analysis for Mendelian disease diagnosis overcomes limitations of conventional genomic testing. J Clin Invest. 2021;131:e141500.
Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–5.
Zhou T, Benda C, Dunzinger S, Huang Y, Ho JC, Yang J, et al. Generation of human induced pluripotent stem cells from urine samples. Nat Protoc. 2012;7:2080–2089.
Xu G, Wu F, Gu X, Zhang J, You K, Chen Y, et al. Direct conversion of human urine cells to neurons by small molecules. Sci Rep. 2019;9:16707.
Loeys BL, Dietz HC, Braverman AC, Callewaert BL, De Backer J, Devereux RB, et al. The revised Ghent nosology for the Marfan syndrome. J Med Genet. 2010;47:476–85.
Akutsu K, Morisaki H, Takeshita S, Ogino H, Higashi M, Okajima T, et al. Characteristics in phenotypic manifestations of genetically proved Marfan syndrome in a Japanese population. Am J Cardiol. 2009;103:1146–8.
Tadaka S, Hishinuma E, Komaki S, Motoike IN, Kawashima J, Saigusa D, et al. jMorp updates in 2020: large enhancement of multi-omics data resources on the general Japanese population. Nucleic Acids Res. 2021;49:D536–D544.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43.
Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018;34:i884–i890.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–31.
Graubert A, Aguet F, Ravi A, Ardlie KG, Getz G. RNA-SeQC 2: Efficient RNA-seq quality control and quantification for large cohorts. Bioinformatics. 2021;37:3048–50.
Yépez VA, Mertes C, Müller MF, Klaproth-Andrade D, Wachutka L, Frésard L, et al. Detection of aberrant gene expression events in RNA sequencing data. Nat Protoc. 2021;16:1276–96.
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6.
Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006;147:126–32.
Wai HA, Lord J, Lyon M, Gunning A, Kelly H, Cibin P, et al. Blood RNA analysis can increase clinical diagnostic rate and resolve variants of uncertain significance. Genet Med. 2020;22:1005–14.
Rahmoune H, Thompson PW, Ward JM, Smith CD, Hong G, Brown J. Glucose transporters in human renal proximal tubular cells isolated from the urine of patients with non-insulin-dependent diabetes. Diabetes. 2005;54:3427–34.
Xue Y, Ankala A, Wilcox WR, Hegde MR. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med. 2015;17:444–51.
Wooderchak-Donahue W, VanSant-Webb C, Tvrdik T, Plant P, Lewis T, Stocks J, et al. Clinical utility of a next generation sequencing panel assay for Marfan and Marfan-like syndromes featuring aortopathy. Am J Med Genet A. 2015;167A:1747–57.
Acknowledgements
We would like to thank the patients for participating in this work. This work was supported by the Japan Agency for Medical Research and Development (AMED) (JP21ek0109549 to T.O.), Grants-in-Aid for Scientific Research (B) (JP20H03641), Grant-in-Aid for Challenging Research (Exploratory) (20K21570) from the Japan Society for the Promotion of Science, Japan Intractable Diseases (Nanbyo) Research Foundation (2020A02), and the Takeda Science Foundation.
Author information
Authors and Affiliations
Contributions
TO and HS contributed to the conception and design of the study. TH, KS, SM, KA, MN, TY, TK, TO, and HS contributed to the acquisition and analysis of data. TH, KS, MN, TO, and HS contributed to drafting the text and preparing the figure. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Hiraide, T., Shimizu, K., Miyamoto, S. et al. Genome sequencing and RNA sequencing of urinary cells reveal an intronic FBN1 variant causing aberrant splicing. J Hum Genet 67, 387–392 (2022). https://doi.org/10.1038/s10038-022-01016-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-022-01016-1
This article is cited by
-
RNA sequencing and target long-read sequencing reveal an intronic transposon insertion causing aberrant splicing
Journal of Human Genetics (2024)
-
A deep intronic TCTN2 variant activating a cryptic exon predicted by SpliceRover in a patient with Joubert syndrome
Journal of Human Genetics (2023)