Abstract
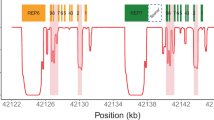
Next-generation sequencing (NGS) has identified variations in cytochrome P450 (CYP) 2D6 associated with drug responses. However, determination of novel haplotypes is difficult because of the short reads generated by NGS. We aimed to identify novel CYP2D6 variants in the Japanese population and predict the CYP2D6 phenotype based on in vitro metabolic studies. Using a targeted NGS panel (PKSeq), 990 Japanese genomes were sequenced, and then novel CYP2D6 haplotypes were determined. Km, Vmax, and intrinsic clearance (Vmax/Km) of N-desmethyl-tamoxifen 4-hydroxylation were calculated by in vitro metabolic studies using cDNA-expressed CYP2D6 proteins. After determination of the CYP2D6 diplotypes, phenotypes of the individuals were predicted based on the in vitro metabolic activities. Targeted NGS identified 14 CYP2D6 variants not registered in the Pharmacogene Variation Consortium (PharmVar) database. Ten novel haplotypes were registered as CYP2D6*128 to *137 alleles in the PharmVar database. Based on the Vmax/Km value of each allele, *128, *129, *130, *131, *132, and *133 were predicted to be nonfunctional alleles. According to the results of the present study, six normal metabolizers (NM) and one intermediate (IM) metabolizers were designated as IM and poor metabolizers (PM), respectively. Our findings provide important insights into novel haplotypes and haplotypes of CYP2D6 and the effects on in vitro metabolic activities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Nofziger C, Turner AJ, Sangkuhl K, Whirl-Carrillo M, Agundez JAG, Black JL, et al. PharmVar GeneFocus: CYP2D6. Clin Pharm Ther. 2020;107:154–70.
Gaedigk A, Ingelman-Sundberg M, Miller NA, Leeder JS, Whirl-Carrillo M, Klein TE, et al. The Pharmacogene Variation (PharmVar) Consortium: incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin Pharm Ther. 2018;103:399–401.
Twist GP, Gaedigk A, Miller NA, Farrow EG, Willig LK, Dinwiddie DL, et al. Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole-genome sequences. NPJ Genom Med. 2016;1:15007.
Gordon AS, Tabor HK, Johnson AD, Snively BM, Assimes TL, Auer PL, et al. Quantifying rare, deleterious variation in 12 human cytochrome P450 drug-metabolism genes in a large-scale exome dataset. Hum Mol Genet. 2014;23:1957–63.
Yang Y, Botton MR, Scott ER, Scott SA. Sequencing the CYP2D6 gene: from variant allele discovery to clinical pharmacogenetic testing. Pharmacogenomics. 2017;18:673–85.
Yoshihama T, Fukunaga K, Hirasawa A, Nomura H, Akahane T, Kataoka F, et al. GSTP1 rs1695 is associated with both hematological toxicity and prognosis of ovarian cancer treated with paclitaxel plus carboplatin combination chemotherapy: a comprehensive analysis using targeted resequencing of 100 pharmacogenes. Oncotarget. 2018;9:29789–800.
Gaedigk A. Complexities of CYP2D6 gene analysis and interpretation. Int Rev Psychiatry. 2013;25:534–53.
Nofziger C, Paulmichl M. Accurately genotyping CYP2D6: not for the faint of heart. Pharmacogenomics. 2018;19:999–1002.
Gaedigk A, Riffel AK, Leeder JS. CYP2D6 haplotype determination using long range allele-specific amplification: resolution of a complex genotype and a discordant genotype involving the CYP2D6*59 allele. J Mol Diagn. 2015;17:740–8.
Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med. 2017;19:69–76.
Hosono N, Kato M, Kiyotani K, Mushiroda T, Takata S, Sato H, et al. CYP2D6 genotyping for functional-gene dosage analysis by allele copy number detection. Clin Chem. 2009;55:1546–54.
Buermans HP, Vossen RH, Anvar SY, Allard WG, Guchelaar HJ, White SJ, et al. Flexible and scalable full-length CYP2D6 long amplicon PacBio sequencing. Hum Mutat. 2017;38:310–6.
Qiao W, Yang Y, Sebra R, Mendiratta G, Gaedigk A, Desnick RJ, et al. Long-read single molecule real-time full gene sequencing of cytochrome P450-2D6. Hum Mutat. 2016;37:315–23.
Fang P, Zheng X, He J, Ge H, Tang P, Cai J, et al. Functional characterization of wild-type and 24 CYP2D6 allelic variants on gefitinib metabolism in vitro. Drug Des Devel Ther. 2017;11:1283–90.
Muroi Y, Saito T, Takahashi M, Sakuyama K, Niinuma Y, Ito M, et al. Functional characterization of wild-type and 49 CYP2D6 allelic variants for N-desmethyltamoxifen 4-hydroxylation activity. Drug Metab Pharmacokinet. 2014;29:360–6.
Kim J, Lim YR, Han S, Han JS, Chun YJ, Yun CH, et al. Functional influence of human CYP2D6 allelic variations: P34S, E418K, S486T, and R296C. Arch Pharm Res. 2013;36:1500–6.
Kiyotani K, Shimizu M, Kumai T, Kamataki T, Kobayashi S, Yamazaki H. Limited effects of frequent CYP2D6*36-*10 tandem duplication allele on in vivo dextromethorphan metabolism in a Japanese population. Eur J Clin Pharm. 2010;66:1065–8.
Caudle KE, Sangkuhl K, Whirl-Carrillo M, Swen JJ, Haidar CE, Klein TE, et al. Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci. 2020;13:116–24.
Momozawa Y, Akiyama M, Kamatani Y, Arakawa S, Yasuda M, Yoshida S, et al. Low-frequency coding variants in CETP and CFB are associated with susceptibility of exudative age-related macular degeneration in the Japanese population. Hum Mol Genet. 2016;25:5027–34.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Hwang S, Kim E, Lee I, Marcotte EM. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep. 2015;5:17875.
Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016;13:1050–54.
Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–100.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Watanabe T, Saito T, Rico EMG, Hishinuma E, Kumondai M, Maekawa M, et al. Functional characterization of 40 CYP2B6 allelic variants by assessing efavirenz 8-hydroxylation. Biochem Pharmacol. 2018;156:420–30.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–8.
Kikura-Hanajiri R, Kawamura M, Miyajima A, Sunouchi M, Goda Y. Chiral analyses of dextromethorphan/levomethorphan and their metabolites in rat and human samples using LC-MS/MS. Anal Bioanal Chem. 2011;400:165–74.
Dahmane E, Mercier T, Zanolari B, Cruchon S, Guignard N, Buclin T, et al. An ultra performance liquid chromatography-tandem MS assay for tamoxifen metabolites profiling in plasma: first evidence of 4’-hydroxylated metabolites in breast cancer patients. J Chromatogr B Anal Technol Biomed Life Sci. 2010;878:3402–14.
Soyama A, Saito Y, Kubo T, Miyajima A, Ohno Y, Komamura K, et al. Sequence-based analysis of the CYP2D6*36-CYP2D6*10 tandem-type arrangement, a major CYP2D6*10 haplotype in the Japanese population. Drug Metab Pharmacokinet. 2006;21:208–16.
Soyama A, Kubo T, Miyajima A, Saito Y, Shiseki K, Komamura K, et al. Novel nonsynonymous single nucleotide polymorphisms in the CYP2D6 gene. Drug Metab Pharmacokinet. 2004;19:313–9.
Higasa K, Miyake N, Yoshimura J, Okamura K, Niihori T, Saitsu H, et al. Human genetic variation database, a reference database of genetic variations in the Japanese population. J Hum Genet. 2016;61:547–53.
Tadaka S, Katsuoka F, Ueki M, Kojima K, Makino S, Saito S, et al. 3.5KJPNv2: an allele frequency panel of 3552 Japanese individuals including the X chromosome. Hum Genome Var. 2019;6:28.
Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19:192–203.
Liau Y, Maggo S, Miller AL, Pearson JF, Kennedy MA, Cree SL. Nanopore sequencing of the pharmacogene CYP2D6 allows simultaneous haplotyping and detection of duplications. Pharmacogenomics. 2019;20:1033–47.
Mantere T, Kersten S, Hoischen A. Long-read sequencing emerging in medical genetics. Front Genet. 2019;10:426.
Ammar R, Paton TA, Torti D, Shlien A, Bader GD. Long read nanopore sequencing for detection of HLA and CYP2D6 variants and haplotypes. F1000Res. 2015;4:17.
Lee SH, Kang S, Dong MS, Park JD, Park J, Rhee S, et al. Characterization of the Ala62Pro polymorphic variant of human cytochrome P450 1A1 using recombinant protein expression. Toxicol Appl Pharm. 2015;285:159–69.
Matsunaga M, Yamazaki H, Kiyotani K, Iwano S, Saruwatari J, Nakagawa K, et al. Two novel CYP2D6*10 haplotypes as possible causes of a poor metabolic phenotype in Japanese. Drug Metab Dispos. 2009;37:699–701.
Aka I, Bernal CJ, Carroll R, Maxwell-Horn A, Oshikoya KA, Van Driest SL. Clinical pharmacogenetics of cytochrome P450-associated drugs in children. J Pers Med. 2017;7:14.
Acknowledgements
We would like to thank Editage (www.editage.com) for English language editing. This study was supported by the Japan Agency for Medical Research and Development (AMED) under Grant Number JP19km0405201 and JP19kk0305009.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Fukunaga, K., Hishinuma, E., Hiratsuka, M. et al. Determination of novel CYP2D6 haplotype using the targeted sequencing followed by the long-read sequencing and the functional characterization in the Japanese population. J Hum Genet 66, 139–149 (2021). https://doi.org/10.1038/s10038-020-0815-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-0815-x
This article is cited by
-
Application of long-read sequencing to elucidate complex pharmacogenomic regions: a proof of principle
The Pharmacogenomics Journal (2022)
-
Methodology for clinical genotyping of CYP2D6 and CYP2C19
Translational Psychiatry (2021)
-
Variation in 100 relevant pharmacogenes among emiratis with insights from understudied populations
Scientific Reports (2020)