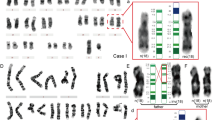
Abstract
Pericentric inversions are among the known polymorphisms detected in the general population at a frequency of 1–2%. Despite their generally benign nature, pericentric inversions affect the reproductive potential of carriers by increasing the risk for unbalanced live-born offspring, miscarriages, or other fertility problems. Here we present a novel large pericentric inversion of chromosome 9, inv(9)(p23q22.3), detected in 30 heterozygote carriers, 24 from seven apparently unrelated families and 6 isolated patients, where the probands were mainly referred for fertility and prenatal problems. The inversion carries a significant risk for recombinant abnormal chromosomes, as in two families one supernumerary rec(9)dup(9p) and one rec(9)dup(9q) were identified, leading to neonatal death and miscarriage, respectively. The inversion carriers were identified by three different laboratories in Greece, Cyprus and Germany respectively, however all carriers have Southeast European origin. The inversion appears to be more frequent in the Greek population, as the majority of the carriers were identified in Greece. We were able to determine that the inversion is identical in all individuals included in the study by applying a combination of several methodologies, such as karyotype, fluorescence in situ hybridization (FISH), chromosomal microarrays (CMA) and haplotype analysis. In addition, haplotype analysis supports that the present inversion is identical by descent (IBD) inherited from a single common ancestor. Our results are, therefore, highly indicative of a founder effect of this inversion, presumably reflecting an event that was present in a small number of individuals that migrated to the current Southeast Europe/Northern Greece from a larger population.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kaiser P. Pericentric inversions. Problems and significance for clinical genetics. Hum Genet. 1984;68:1–47.
Kosyakova N, Grigorian A, Liehr T, Manvelyan M, Simonyan I, Mkrtchyan H, et al. Heteromorphic variants of chromosome 9. Mol Cytogenet. 2013;6:14.
Mattei JF, Mattei MG, Ardissone JP, Taramasco H, Giraud F. Pericentric inversion, inv(9) (p22 q32), in the father of a child with a duplication-deletion of chromosome 9 and gene dosage effect for adenylate kinase-1. Clin Genet. 1980;17:129–36.
Mattei JF, Mattei MG, Balestrazzi P, Giraud F. Familial pericentric inversion of chromosome 9, INV(9)(p22q32) with recurrent duplication-deletion. Clin Genet. 1983;24:220–2.
Shapira SK, Orr-Urtreger A, Gagos S, Shaffer LG. Constitutional mosaicism for a chromosome 9 inversion resulting in recombinant aneusomy in an offspring. Am J Med Genet. 1997;69:360–4.
Sonoda T, Ohba K, Ohdo S, Sameshima K. 9p deletion and distal 9q duplication due to a paternal pericentric inversion 9(p22q32). Jpn J Hum Genet. 1991;36:111–6.
Mundhofir FE, Smeets D, Nillesen W, Winarni TI, Yntema HG, de Leeuw N, et al. Monosomy 9pter and trisomy 9q34.11qter in two sisters due to a maternal pericentric inversion. Gene. 2012;511:451–4.
Gardner RJM, Amor DJ. Gardner and Sutherland’s chromosome abnormalities and genetic counseling. 5 ed. Oxford University Press; Oxford, 2018.
Kurahashi H, Emanuel BS. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet. 2001;10:2605–17.
Fickelscher I, Liehr T, Watts K, Bryant V, Barber JC, Heidemann S, et al. The variant inv(2)(p11.2q13) is a genuinely recurrent rearrangement but displays some breakpoint heterogeneity. Am J Hum Genet. 2007;81:847–56.
Gilling M, Dullinger JS, Gesk S, Metzke-Heidemann S, Siebert R, Meyer T, et al. Breakpoint cloning and haplotype analysis indicate a single origin of the common Inv(10)(p11.2q21.2) mutation among northern Europeans. Am J Hum Genet. 2006;78:878–83.
Thomas NS, Bryant V, Maloney V, Cockwell AE, Jacobs PA. Investigation of the origins of human autosomal inversions. Hum Genet. 2008;123:607–16.
Liehr TC, Claussen U. Multicolor-FISH approaches for the characterization of human chromosomes in clinical genetics and tumor cytogenetics. Curr Genom.2002;3:213–35.
Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–4.
Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89.
Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–62.
Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–21.
MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–92.
Martinez-Fundichely A, Casillas S, Egea R, Ramia M, Barbadilla A, Pantano L, et al. InvFEST, a database integrating information of polymorphic inversions in the human genome. Nucleic Acids Res. 2014;42:D1027–32.
Lee J, Han K, Meyer TJ, Kim HS, Batzer MA. Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons. PloS One. 2008;3:e4047.
Salm MP, Horswell SD, Hutchison CE, Speedy HE, Yang X, Liang L, et al. The origin, global distribution, and functional impact of the human 8p23 inversion polymorphism. Genom Res. 2012;22:1144–53.
Ma J, Amos CI. Investigation of inversion polymorphisms in the human genome using principal components analysis. PloS One. 2012;7:e40224.
Garcia-Peiro A, Oliver-Bonet M, Navarro J, Abad C, Guitart M, Amengual MJ, et al. Dynamics of sperm DNA fragmentation in patients carrying structurally rearranged chromosomes. Int J Androl. 2011;34(6 Pt 2):e546–53.
Serra A, Brahe C, Millington-Ward A, Neri G, Tedeschi B, Tassone F, et al. Pericentric inversion of chromosome 9: prevalence in 300 Down syndrome families and molecular studies of nondisjunction. Am J Med Genet Suppl. 1990;7:162–8.
Puig M, Casillas S, Villatoro S, Caceres M. Human inversions and their functional consequences. Brief Funct Genom. 2015;14:369–79.
Author information
Authors and Affiliations
Contributions
VV conceived and designed the project. VV and CS prepared the manuscript. CS and SMR contributed equally in the design and execution of the experiments. SMR contributed in the final preparation of the manuscript. SF contributed in the final corrections of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics
This study was approved by the scientific committee of Bioiatriki Healthcare Group and all participants provided written informed consent prior to testing and participation.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Sismani, C., Rapti, SM., Iliopoulou, P. et al. Novel pericentric inversion inv(9)(p23q22.3) in unrelated individuals with fertility problems in the Southeast European population. J Hum Genet 65, 783–795 (2020). https://doi.org/10.1038/s10038-020-0769-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-0769-z
This article is cited by
-
The Relationship Between Chromosomal Polymorphism and Male Reproductive Abnormalities
Reproductive Sciences (2024)