Abstract
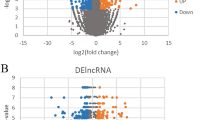
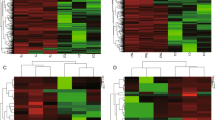
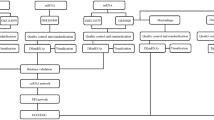
The metabolic syndrome (MS) is a cluster of interrelated risk factors including diabetes mellitus, abdominal obesity, high cholesterol, and hypertension, which can significantly increase mortality and disability. Accumulating evidence suggest that long non-coding RNAs (lncRNAs) are involved in the pathogenesis of human metabolic diseases. However, little is known about the regulatory role of lncRNAs in MS. In this work, we proposed a method for identifying potential MS-associated lncRNAs by constructing an lncRNA–miRNA–mRNA network (LMMN). Firstly, we constructed LMMN by integrating MS-associated genes, miRNA–mRNA interactions, miRNA–lncRNA interactions and mRNA/miRNA expression profiles in patients with MS. Then, we predicted potential MS-associated lncRNAs based on the topological properties of LMMN. As a result, we identified XIST as the most important lncRNA in LMMN. Furthermore, we focused on XIST/miR-214-3p and mir-181a-5p/PTEN axis and validated their expression in MS using real-time quantitative polymerase chain reaction (RT-qPCR). The RT-qPCR results showed that the expression of XIST and PTEN was significantly decreased (P < 0.05) while the expression of miR-214-3p was significantly increased (P < 0.05) in peripheral blood mononuclear cells (PBMCs) of patients with MS, compared with healthy controls. In addition, correlation analysis showed that XIST was negatively correlated with serum C peptide and PTEN was positively correlated with BMI of MS patients. Our findings provided new evidence for further exploring the regulatory role of XIST and other lncRNAs in MS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Users can freely access MS-associated lncRNA–miRNA–mRNA network, XIST-centered lncRNA–miRNA–mRNA network and XIST–miRNA–PTEN network through the following link: http://www.ndexbio.org/#/networkset/df19b350-593a-11ea-bfdc-0ac135e8bacf?accesskey=771d8c38dc933a19f1ccbfb5b1c3c9105177b8170879bd4215c1d0a6ec62cd16.
References
Hudish LI, Reusch JE, Sussel L. beta Cell dysfunction during progression of metabolic syndrome to type 2 diabetes. J Clin Investig. 2019;129:4001–8.
Mottillo S, Filion KB, Genest J, Joseph L, Pilote L, Poirier P, et al. The metabolic syndrome and cardiovascular risk: a systematic review and meta-analysis. J Am Coll Cardiol. 2010;56:1113–32.
Baxter AJ, Coyne T, McClintock C. Dietary patterns and metabolic syndrome-a review of epidemiologic evidence. Asia Pac J Clin Nutr. 2006;15:134–42.
Cho N, Shaw JE, Karuranga S, Huang Y, da Rocha Fernandes JD, Ohlrogge AW, et al. IDF Diabetes Atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract. 2018;138:271–81.
Sun J, Ruan Y, Wang M, Chen R, Yu N, Sun L, et al. Differentially expressed circulating LncRNAs and mRNA identified by microarray analysis in obese patients. Sci Rep. 2016;6:35421.
Nuermaimaiti N, Liu J, Liang X, Jiao Y, Zhang D, Liu L, et al. Effect of lncRNA HOXA11-AS1 on adipocyte differentiation in human adipose-derived stem cells. Biochem Biophys Res Commun. 2018;495:1878–84.
Ruan Y, Lin N, Ma Q, Chen R, Zhang Z, Wen W, et al. Circulating LncRNAs analysis in patients with type 2 diabetes reveals novel genes influencing glucose metabolism and Islet β-cell function. Cell Physiol Biochem. 2018;46:335–50.
Zhu X, Wu YB, Zhou J, Kang DM. Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via increasing FoxO1 expression. Biochem Biophys Res Commun. 2016;469:319–25.
Shi L, Tian C, Sun L, Cao F, Meng Z. The lncRNA TUG1/miR-145-5p/FGF10 regulates proliferation and migration in VSMCs of hypertension. Biochem Biophys Res Commun. 2018;501:688–95.
Losko M, Kotlinowski J, Jura J. Long noncoding RNAs in metabolic syndrome related disorders. Mediators Inflamm. 2016.
Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307.
Sathishkumar C, Prabu P, Mohan V, Balasubramanyam M. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum Genomics. 2018;12:41.
Liu HZ, Wang QY, Zhang Y, Qi DT, Li MW, Guo WQ, et al. Pioglitazone up-regulates long non-coding RNA MEG3 to protect endothelial progenitor cells via increasing HDAC7 expression in metabolic syndrome. Biomed Pharmacother. 2016;78:101–9.
Chen X, Yan CC, Zhang X, You ZH. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2017;18:558–76.
Dai HJ, Wu JCY, Tsai RTH, Pan WH, Hsu WL. T-HOD: a literature-based candidate gene database for hypertension, obesity and diabetes. Database. 2013;bas061.
Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–D97.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011;146:353–8.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–4.
D’Amore S, Härdfeldt J, Cariello M, Graziano G, Copetti M, Di Tullio G, et al. Identification of miR-9-5p as direct regulator of ABCA1 and HDL-driven reverse cholesterol transport in circulating CD14+ cells of patients with metabolic syndrome. Cardiovasc Res. 2018;114:1154–64.
Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–7.
Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA and Dudoit S, editors. Bioinformatics and computational biology solutions using R and bioconductor. New York, NY: Springer; 2005. p. 397–420.
Chen D, Lü L, Shang MS, Zhang YC, Zhou T. Identifying influential nodes in complex networks. Physica A. 2012;391:1777–87.
Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419–D426.
Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47(D1):D590–D595.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Pal A, Barber TM, Van de Bunt M, Rudge SA, Zhang Q, Lachlan KL, et al. PTEN mutations as a cause of constitutive insulin sensitivity and obesity. N Engl J Med. 2012;367:1002–11.
Gao JR, Qin XJ, Fang ZH, Han LP, Guo MF, Jiang NN. To explore the pathogenesis of vascular lesion of type 2 diabetes mellitus based on the PI3K/Akt signaling pathway. J Diabetes Res. 2019;2019:4650906. https://doi.org/10.1155/2019/4650906.
Zhao XC, Yang SH, Yan YQ, Zhang X, Zhang L, Jiao B, et al. Identification of differential gene expression profile from peripheral blood cells of military pilots with hypertension by RNA sequencing analysis. BMC Med Genom. 2018;11:59.
Chen H, Fajol A, Hoene M, Zhang B, Schleicher ED, Lin Y, et al. PI3K-resistant GSK3 controls adiponectin formation and protects from metabolic syndrome. Proc Natl Acad Sci USA. 2016;113:5754–9.
Lozano-Bartolomé J, Llauradó G, Portero-Otin M, Altuna-Coy A, Rojo-Martínez G, Vendrell J, et al. Altered expression of miR-181a-5p and miR-23a-3p is associated with obesity and TNF α-induced insulin resistance. J Clin Endocrinol Metab. 2018;103:1447–58.
Honardoost M, Keramati F, Arefian E, Mohammadi Yeganeh S, Soleimani M. Network of three specific microRNAs influence type 2 diabetes through inducing insulin resistance in muscle cell lines. J Cell Biochem. 2019;120:1532–8.
Hulsmans M, Sinnaeve P, Van der Schueren B, Mathieu C, Janssens S, Holvoet P. Decreased miR-181a expression in monocytes of obese patients is associated with the occurrence of metabolic syndrome and coronary artery disease. J Clin Endocrinol Metab. 2012;97:E1213–E1218.
Kitamoto A, Kitamoto T, Mizusawa S, Teranishi H, So R, Matsuo T, et al. NUDT3 rs206936 is associated with body mass index in obese Japanese women. Endocr J. 2013;60:991–1000.
Matsuda S, Kobayashi M, Kitagishi Y. Roles for PI3K/AKT/PTEN pathway in cell signaling of nonalcoholic fatty liver disease. ISRN Endocrinol. 2013;2013:1–7.
Ramzan F, D’Souza RF, Durainayagam BR, Milan AM, Markworth JF, Miranda-Soberanis V, et al. Circulatory miRNA biomarkers of metabolic syndrome. Acta Diabetol. 2020;57:203–14.
Ma E, Fu Y, Garvey WT. Relationship of circulating miRNAs with insulin sensitivity and associated metabolic risk factors in humans. Metab Syndr Relat Disord. 2018;16:82–9.
Kim HJ, Kobayashi M, Sasaki T, Kikuchi O, Amano K, Kitazumi T, et al. Overexpression of FoxO1 in the hypothalamus and pancreas causes obesity and glucose intolerance. Endocrinology. 2012;153:659–71.
Tonks KT, Ng Y, Miller S, Coster AC, Samocha-Bonet D, Iseli TJ, et al. Impaired Akt phosphorylation in insulin-resistant human muscle is accompanied by selective and heterogeneous downstream defects. Diabetologia. 2013;56:875–85.
Battelli MG, Bortolotti M, Polito L, Bolognesi A. Metabolic syndrome and cancer risk: the role of xanthine oxidoreductase. Redox Biol. 2019;21:101070.
Chang S, Chen B, Wang X, Wu K, Sun Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017;17:248.
Liu H, Yin T, Yan W, Si R, Wang B, Chen M, et al. Dysregulation of microRNA-214 and PTEN contributes to the pathogenesis of hypoxic pulmonary hypertension. Int J Chron Obstruct Pulmon Dis. 2017;12:1781.
Wang C, Qi S, Xie C, Li C, Wang P, Liu D. Upregulation of long non-coding RNA XIST has anticancer effects on epithelial ovarian cancer cells through inverse downregulation of hsa-miR-214-3p. J Gynecol Oncol. 2018;29:e99. https://doi.org/10.3802/jgo.2018.29.e99.
Geletina NS, Kobelev VS, Babayants EV, Feng L, Pustylnyak VO, Gulyaeva LF. PTEN negative correlates with miR-181a in tumour tissues of non-obese endometrial cancer patients. Gene. 2018;655:20–4.
Li K, Zhang J, Yu J, Liu B, Guo Y, Deng J, et al. MicroRNA-214 suppresses gluconeogenesis by targeting activating transcriptional factor 4. J Biol Chem. 2015;290:8185–95.
Cui X, Tan J, Shi Y, Sun C, Li Y, Ji C, et al. The long non-coding RNA Gm10768 activates hepatic gluconeogenesis by sequestering microRNA-214 in mice. J Biol Chem. 2018;293:4097–109.
Yan Z, Zang B, Gong X, Ren J, Wang R. MiR-214-3p exacerbates kidney damages and inflammation induced by hyperlipidemic pancreatitis complicated with acute renal injury. Life Sci. 2020;241:117118.
Alexandru N, Constantin A, Nemecz M, Comariţa IK, Vîlcu A, Procopciuc A, et al. Hypertension associated with hyperlipidemia induced different microRNA expression profiles in plasma, platelets, and platelet-derived microvesicles; effects of endothelial progenitor cell therapy. Front Med (Lausanne). 2019;6:280. https://doi.org/10.3389/fmed.2019.00280.
Acknowledgements
This work was supported by Innovation Talents Project of Harbin Science and Technology Bureau (2017RAQXJ027), the Natural Science Foundation of Heilongjiang Province (LH2019F023), the Fundamental Research Foundation for Universities of Heilongjiang Province (LGYC2018JQ003), and China Scholarship Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yao, D., Lin, Z., Zhan, X. et al. Identifying potential functional lncRNAs in metabolic syndrome by constructing a lncRNA–miRNA–mRNA network. J Hum Genet 65, 927–938 (2020). https://doi.org/10.1038/s10038-020-0753-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-0753-7