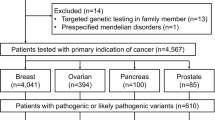
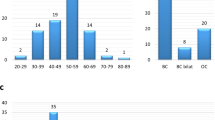
Abstract
Genetic testing for BRCA1/2 mutations has become the standard clinical practice. Recent findings suggest the clinical significance of multigene panel testing of BRCA1/2 and other cancer-related genes. However, the clinical features of patients with breast cancer with germline mutations identified using multigene panels remain unclear. In this study, DNA samples from 583 Chinese women with breast cancer were subjected to target sequencing for 54 cancer-related genes using a pre-capture pooling method followed by next-generation sequencing. We identified 79 pathogenic germline mutations in 21 cancer-related genes. Forty-five patients (7.7%) harbored BRCA1/2 mutations, and 38 patients (6.5%) carried pathogenic mutations in the remaining 19 genes. PALB2 was the most commonly (1.2%) mutated gene other than BRCA1/2. Most of the identified pathogenic mutations were novel, suggesting mutation screening by using multigene panel testing is important particularly for non-European populations. Mutations in BRCA1/2 and the other cancer-related genes were differentially associated with clinical features. BRCA1 mutation carriers were strongly associated with triple-negative breast cancer (TNBC), whereas BRCA2 mutation carriers were not. Tumors in BRCA1-mutation carriers had a high histological grade. Patients with BRCA2-mutated breast cancers were likely to develop E-cadherin-negative tumors with bone metastases. Furthermore, mutations in PALB2 were strongly associated with TNBC. We demonstrated the usefulness of multigene panel testing and observed that a substantial proportion of patients with breast cancer had hereditary risk factors. Identifying differential associations between mutation status and clinical features will advance our understanding regarding the pathologies of this heterogeneous disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67:439–48.
Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40:17–22.
Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7:a016600.
King MC. “The race” to clone BRCA1. Science. 2014;343:1462–5.
King MC, Marks JH, Mandell JB, New York Breast Cancer Study Group. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–6.
Wallace AJ. New challenges for BRCA testing: a view from the diagnostic laboratory. Eur J Hum Genet. 2016;24(Suppl 1):S10–8.
Easton DF, Pharoah PD, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N. Engl J Med. 2015;372:2243–57.
Malkin D, Li FP, Strong LC, Fraumeni JF, Nelson CE, Kim DH, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–8.
Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–7.
Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7.
Antoniou AC, Foulkes WD, Tischkowitz M. Breast-cancer risk in families with mutations in PALB2. N. Engl J Med. 2014;371:1651–2.
Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R. et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–9.
Weischer M, Bojesen SE, Ellervik C, Tybjaerg-Hansen A, Nordestgaard BG. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol. 2008;26:542–8.
Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–5.
Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–51.
Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ, et al. Strategies for subtypes-dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–47.
Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–90.
Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16:110–20.
Ahmadloo S, Nakaoka H, Hayano T, Hosomichi K, You H, Utsuno E, et al. Rapid and cost-effective high-throughput sequencing for identification of germline mutations of BRCA1 and BRCA2. J Hum Genet. 2017;62:561–7.
Zheng K, Tan JX, Li F, Li HY, Zeng XH, Ma BL, et al. Clinicopathologic factors related to the histological tumor grade of breast cancer in Western China: an epidemiological multicenter study of 8619 female patients. Transl Oncol. 2018;11:1023–33.
Suda K, Nakaoka H, Yoshihara K, Ishiguro T, Tamura R, Mori Y, et al. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep. 2018;24:1777–89.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–8.
Hata C, Nakaoka H, Xiang Y, Wang D, Yang A, Liu D, et al. Exploration of intermediate-sized INDELs by next-generation multigene panel testing in Han Chinese patients with breast cancer. Hum Genome Var. 2019;6:51.
Houdayer C, Caux-Moncoutier V, Krieger S, Barrois M, Bonnet F, Bourdon V, et al. Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat. 2012;33:1228–38.
Kwong A, Shin VY, Ho JC, Kang E, Nakamura S, Teo SH, et al. Comprehensive spectrum of BRCA1 and BRCA2 deleterious mutations in breast cancer in Asian countries. J Med Genet. 2016;53:15–23.
Foulkes WD, Stefansson IM, Chappuis PO, Bégin LR, Goffin JR, Wong N, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95:1482–5.
Laakso M, Loman N, Borg A, Isola J. Cytokeratin 5/14-positive breast cancer: true basal phenotype confined to BRCA1 tumors. Mod Pathol. 2005;18:1321–8.
NCCN Guidelines. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Genetic/Familial High-Risk Assessment: Breast and Ovarian. (2017, October 3). Available at https://www.genomeweb.com/sites/default/files/nccn_2017.pdf. Accessed November 10, 2019.
Sun J, Meng H, Yao L, Lv M, Bai J, Zhang J, et al. Germline mutations in cancer susceptibility genes in a large series of unselected breast cancer patients. Clin Cancer Res. 2017;23:6113–9.
Momozawa Y, Iwasaki Y, Parsons MT, Kamatani Y, Takahashi A, Tamura C, et al. Germline pathogenic variants of 11 breast cancer genes in 7,051 Japanese patients and 11,241 controls. Nat Commun. 2018;9:4083.
Couch FJ, Shimelis H, Hu C, Hart SN, Polley EC, Na J, et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017;3:1190–6.
Bogdanova N, Cybulski C, Bermisheva M, Datsyuk I, Yamini P, Hillemanns P, et al. A nonsense mutation (E1978X) in the ATM gene is associated with breast cancer. Breast Cancer Res Treat. 2009;118:207–11.
Cybulski C, Wokołorczyk D, Jakubowska A, Huzarski T, Byrski T, Gronwald J, et al. Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer. J Clin Oncol. 2011;29:3747–52.
Warner E, Foulkes W, Goodwin P, Meschino W, Blondal J, Paterson C, et al. Prevalence and penetrance of BRCA1 and BRCA2 gene mutations in unselected Ashkenazi Jewish women with breast cancer. J Natl Cancer Inst. 1999;91:1241–7.
Frey MK, Kopparam RV, Ni Zhou Z, Fields JC, Buskwofie A, Carlson AD, et al. Prevalence of nonfounder BRCA1/2 mutations in Ashkenazi Jewish patients presenting for genetic testing at a hereditary breast and ovarian cancer center. Cancer. 2019;125:690–7.
Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–30.
Chiang CWK, Mangul S, Robles C, Sankararaman S. A comprehensive map of genetic variation in the world’s largest ethnic group-Han Chinese. Mol Biol Evol. 2018;35:2736–50.
Qureshi HS, Linden MD, Divine G, Raju UB. E-cadherin status in breast cancer correlates with histologic type but does not correlate with established prognostic parameters. Am J Clin Pathol. 2006;125:377–85.
Shimelis H, LaDuca H, Hu C, Hart SN, Na J, Thomas A, et al. Triple-negative breast cancer risk genes identified by multigene hereditary cancer panel testing. J Natl Cancer Inst. 2018;110:855–62.
Sunada S, Nakanishi A, Miki Y. Crosstalk of DNA double-strand break repair pathways in poly(ADP-ribose) polymerase inhibitor treatment of breast cancer susceptibility gene 1/2-mutated cancer. Cancer Sci. 2018;109:893–9.
Stevens KN, Vachon CM, Couch FJ. Genetic susceptibility to triple-negative breast cancer. Cancer Res. 2013;73:2025–30.
Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11:5175–80.
Woods NT, Baskin R, Golubeva V, Jhuraney A, De-Gregoriis G, Vaclova T, et al. Functional assays provide a robust tool for the clinical annotation of genetic variants of uncertain significance. NPJ Genom Med. 2016;1:16001.
Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–22.
Starita LM, Islam MM, Banerjee T, Adamovich AI, Gullingsrud J, Fields S, et al. A multiplex homology-directed DNA repair assay reveals the impact of more than 1,000 BRCA1 missense substitution variants on protein function. Am J Hum Genet. 2018;103:498–508.
Matreyek KA, Starita LM, Stephany JJ, Martin B, Chiasson MA, Gray VE, et al. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat Genet. 2018;50:874–82.
Acknowledgements
We are grateful to the participants in this study. The authors would like to thank Enago (www.enago.jp) for the English language review. This project was mainly supported by Guangzhou Medical University and National Institute of Genetics (NIG Collaborative Research Program). Hua You is supported by the National Natural Science Foundation of China (81911530169, 81903088, 81670180, 81711540047, and 81850410547). Li Wei is supported by Venture & Innovation Support Program for Chongqing Overseas Returnees (cx2019051). Ke Zheng is supported by the National Natural Science Foundation of China (81202090). Dahai Liu is supported by the National Natural Science Foundation of China (81870307), the University Special Innovative Research Program of Department of Education of Guangdong Province (2017KTSCX189). Ituro Inoue is supported by JSPS and NSFC under the Japan-China Scientific Cooperation Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hata, C., Nakaoka, H., Xiang, Y. et al. Germline mutations of multiple breast cancer-related genes are differentially associated with triple-negative breast cancers and prognostic factors. J Hum Genet 65, 577–587 (2020). https://doi.org/10.1038/s10038-020-0729-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-0729-7
This article is cited by
-
A systematic review of the prevalence of germline pathogenic variants in patients with pancreatic cancer
Journal of Gastroenterology (2021)
-
Assessment of tumor suppressor promoter methylation in healthy individuals
Clinical Epigenetics (2020)
-
A commentary on germline mutations of multiple breast cancer-related genes are differentially associated with triple-negative breast cancers and prognostic factors
Journal of Human Genetics (2020)