Abstract
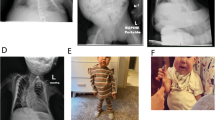
Kagami–Ogata syndrome (KOS14) is a rare imprinting disorder characterized by a unique constellation of phenotypes including bell-shaped small thorax with coat-hanger appearance of the ribs. We encountered an African American female infant with KOS14 phenotype and 46,XX,t(2;14)(q11.2;q32.2)mat. After excluding upd(14)pat and an epimutation (hypermethylation) and a deletion affecting the maternally derived 14q32.2 imprinted region, we performed whole-genome sequencing, revealing that the translocation was generated between noncoding region at 2q11.2 and intron 6 of MEG3 at 14q32.2. Subsequent Sanger sequencing for the fusion points showed that the chromosomal fusion on the der(2) chromosome occurred between Chr2:102,193,994 (bp) and Chr14:101,314,628 (bp) in association with an insertion of 5-bp segment of unknown origin and that on the der(14) chromosome took place between Chr14:101,314,627 (bp) and Chr2:102,193,995 (bp) in association with an insertion of 1-bp segment of unknown origin (according to GRCh37/hg19). The results, together with the previous data in patients with KOS14, imply that the MEG3 disruption by 46,XX,t(2;14)(q11.2;q32.2)mat caused silencing of all MEGs including RTL1as and resultant excessive RTL1 expression, leading to the development of KOS14. To our knowledge, while Robertsonian translocations involving chromosome 14 have been reported in KOS14, this is the first case of KOS14 caused by a chromosomal translocation involving the 14q32.2 imprinted region.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ogata T, Kagami M. Kagami-Ogata syndrome: a clinically recognizable upd(14)pat and related disorder affecting the chromosome 14q32.2 imprinted region. J Hum Genet. 2016;61:87–94.
Kagami M, Sekita Y, Nishimura G, Irie M, Kato F, Okada M, et al. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet. 2008;40:237–42.
Kagami M, Matsuoka K, Nagai T, Yamanaka M, Kurosawa K, Suzumori N, et al. Paternal uniparental disomy 14 and related disorders: placental gene expression analyses and histological examinations. Epigenetics. 2012;7:1142–50.
Kagami M, O’Sullivan MJ, Green AJ, Watabe Y, Arisaka O, Masawa N, et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet. 2010;6:e1000992.
Kagami M, Kurosawa K, Miyazaki O, Ishino F, Matsuoka K, Ogata T. Comprehensive clinical studies in 34 patients with molecularly defined UPD(14)pat and related conditions (Kagami-Ogata syndrome). Eur J Hum Genet. 2015;23:1488–98.
Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–2.
Suzuki T, Tsurusaki Y, Nakashima M, Miyake N, Saitsu H, Takeda S, et al. Precise detection of chromosomal translocation or inversion breakpoints by whole-genome sequencing. J Hum Genet. 2014;59:649–54.
Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4.
Tierling S, Dalbert S, Schoppenhorst S, Tsai CE, Oliger S, Ferguson-Smith AC, et al. High-resolution map and imprinting analysis of the Gtl2-Dnchc1 domain on mouse chromosome 12. Genomics. 2007;87:225–35.
da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–16.
Acknowledgements
We thank Ms Aya Kitamoto and Mr Naoki Adachi for their technical support.
Funding
This study was funded by Japan Agency for Medical Research and Development (AMED) (JP19ek0109301 to TO and JP19ek0109373 to MK).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Omark, J., Masunaga, Y., Hannibal, M. et al. Kagami–Ogata syndrome in a patient with 46,XX,t(2;14)(q11.2;q32.2)mat disrupting MEG3. J Hum Genet 66, 439–443 (2021). https://doi.org/10.1038/s10038-020-00858-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-00858-x