Abstract
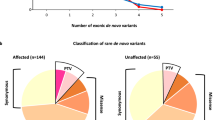
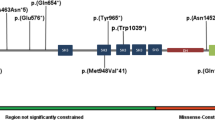
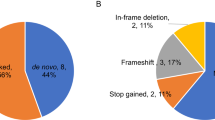
Autism spectrum disorder (ASD) is a complex neurodevelopmental disorder with heterogeneity in presentation, genetic etiology, and clinical outcome. Although numerous ASD susceptibility genes have been described, they only account for a small fraction of the estimated heritability, supporting the need to identify more risk variants. This study reports the whole exome sequencing for 24 simplex families with sporadic cases of ASD. These families were selected following a rigorous family history study designed to exclude families with any history of neurodevelopmental or psychiatric disease. Fifteen rare, de novo variants, including fourteen missense variants and one splicing variant, in thirteen families were identified. We describe a splicing variant in XRCC6 which was predicted to destroy the 5′ splice site in intron 9 and introduce a premature stop codon. We observed intron 9 retention in XRCC6 transcripts and reduced XRCC6 expression in the proband. Reduced XRCC6 activity and function may be relevant to ASD etiology due to XRCC6’s role in nonhomologous DNA repair and interactions of the C-terminal SAP domain with DEAF1, a nuclear transcriptional regulator that is important during embryonic development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. J Am Med Assoc. 2015;314:895–903.
Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci. 2015;16:551–63.
Masi A, DeMayo MM, Glozier N, Guastella AJ. An overview of autism spectrum disorder, heterogeneity and treatment options. Neurosci Bull. 2017;33:183–93.
Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77.
Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–94.
Ramaswami G, Geschwind DH. Genetics of autism spectrum disorder. Handb Clin Neurol. 2018;147:321–9.
Balicza P, Varga NA, Bolgar B, Pentelenyi K, Bencsik R, Gal A, et al. Comprehensive analysis of rare variants of 101 autism-linked genes in a hungarian cohort of autism spectrum disorder patients. Front Genet. 2019;10:434.
SPARK. A US cohort of 50,000 families to accelerate autism research. Neuron. 2018;97:488–93.
Jiang YH, Yuen RK, Jin X, Wang M, Chen N, Wu X, et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013;93:249–63.
De Rubeis S, Buxbaum JD. Genetics and genomics of autism spectrum disorder: embracing complexity. Hum Mol Genet. 2015;24:R24–R31.
O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–9.
Feliciano P, Zhou X, Astrovskaya I, Turner TN, Wang T, Brueggeman L, et al. Exome sequencing of 457 autism families recruited online provides evidence for autism risk genes. NPJ Genom Med. 2019;4:19.
Yuen RKC, Merico D, Bookman M, Howe L, Thiruvahindrapuram B, Patel RV, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci. 2017;20:602–11.
Ruzzo EK, Perez-Cano L, Jung JY, Wang LK, Kashef-Haghighi D, Hartl C, et al. Inherited and de novo genetic risk for autism impacts shared networks. Cell. 2019;178:850–66.
Turner TN, Coe BP, Dickel DE, Hoekzema K, Nelson BJ, Zody MC, et al. Genomic patterns of de novo mutation in simplex autism. Cell. 2017;171:710–22 e12.
Yoo H. Genetics of autism spectrum disorder: current status and possible clinical applications. Exp Neurobiol. 2015;24:257–72.
Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4:a009886.
Doherty JL, Owen MJ. Genomic insights into the overlap between psychiatric disorders: implications for research and clinical practice. Genome Med. 2014;6:29.
Carroll LS, Owen MJ. Genetic overlap between autism, schizophrenia and bipolar disorder. Genome Med. 2009;1:102.
Cohen IL, Schmidt-Lackner S, Romanczyk R, Sudhalter V. The PDD behavior inventory: a rating scale for assessing response to intervention in children with pervasive developmental disorder. J Autism Dev Disord. 2003;33:31–45.
Cohen IL, Liu X, Hudson M, Gillis J, Cavalari RN, Romanczyk RG, et al. Using the PDD behavior inventory as a Level 2 Screener: a classification and regression trees analysis. J Autism Dev Disord. 2016;46:3006–22.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Exome Variant Server. NHLBI GO exome sequencing project (ESP). Seattle, WA: Exome Variant Server; 2017 http://evs.gs.washington.edu/EVS/.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–74.
Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4.
Reva B, Antipin Y, Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 2007;8:R232.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–73.
Douville C, Carter H, Kim R, Niknafs N, Diekhans M, Stenson PD, et al. CRAVAT: cancer-related analysis of variants toolkit. Bioinformatics. 2013;29:647–8.
Masica DL, Douville C, Tokheim C, Bhattacharya R, Kim R, Moad K, et al. CRAVAT 4: cancer-related analysis of variants toolkit. Cancer Res. 2017;77:e35–e8.
Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet. 2009;84:524–33.
Zeisel A, Yitzhaky A, Bossel Ben-Moshe N, Domany E. An accessible database for mouse and human whole transcriptome qPCR primers. Bioinformatics. 2013;29:1355–6.
Hultman CM, Sandin S, Levine SZ, Lichtenstein P, Reichenberg A. Advancing paternal age and risk of autism: new evidence from a population-based study and a meta-analysis of epidemiological studies. Mol Psychiatry. 2011;16:1203–12.
O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50.
Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67.
Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci USA. 2010;107:961–8.
Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–14.
Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–4.
Jensik PJ, Huggenvik JI, Collard MW. Deformed epidermal autoregulatory factor-1 (DEAF1) interacts with the Ku70 subunit of the DNA-dependent protein kinase complex. PLoS One. 2012;7:e33404.
Vulto-van Silfhout AT, Rajamanickam S, Jensik PJ, Vergult S, de RN, Newhall KJ, et al. Mutations affecting the SAND domain of DEAF1 cause intellectual disability with severe speech impairment and behavioral problems. Am J Hum Genet. 2014;94:649–61.
Chen L, Jensik PJ, Alaimo JT, Walkiewicz M, Berger S, Roeder E, et al. Functional analysis of novel DEAF1 variants identified through clinical exome sequencing expands DEAF1-associated neurodevelopmental disorder (DAND) phenotype. Hum Mutat. 2017;38:1774–85.
Casanova EL, Sharp JL, Chakraborty H, Sumi NS, Casanova MF. Genes with high penetrance for syndromic and non-syndromic autism typically function within the nucleus and regulate gene expression. Mol Autism. 2016;7:18.
Rajab A, Schuelke M, Gill E, Zwirner A, Seifert F, Morales GS, et al. Recessive DEAF1 mutation associates with autism, intellectual disability, basal ganglia dysfunction and epilepsy. J Med Genet. 2015;52:607–11.
Miller JN, Pearce DA. Nonsense-mediated decay in genetic disease: friend or foe? Mutat Res Rev Mutat Res. 2014;762:52–64.
Bladen CL, Navarre S, Dynan WS, Kozlowski DJ. Expression of the Ku70 subunit (XRCC6) and protection from low dose ionizing radiation during zebrafish embryogenesis. Neurosci Lett. 2007;422:97–102.
Yu W, Li L, Wang G, Zhang W, Xu J, Liang A. KU70 inhibition impairs both non-homologous end joining and homologous recombination DNA damage repair through SHP-1 induced dephosphorylation of SIRT1 in T-cell acute lymphoblastic leukemia (T-ALL) [corrected]. Cell Physiol Biochem. 2018;49:2111–23.
Kassam I, Wu Y, Yang J, Visscher PM, McRae AF. Tissue-specific sex differences in human gene expression. Hum Mol Genet. 2019;28:2976–86.
Zang Y, Pascal LE, Zhou Y, Qiu X, Wei L, Ai J, et al. ELL2 regulates DNA non-homologous end joining (NHEJ) repair in prostate cancer cells. Cancer Lett. 2018;415:198–207.
Jimenez-Barron LT, O'Rawe JA, Wu Y, Yoon M, Fang H, Iossifov I, et al. Genome-wide variant analysis of simplex autism families with an integrative clinical-bioinformatics pipeline. Cold Spring Harb Mol Case Stud. 2015;1:a000422.
Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21.
Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41.
Cheaha D, Bumrungsri S, Chatpun S, Kumarnsit E. Characterization of in utero valproic acid mouse model of autism by local field potential in the hippocampus and the olfactory bulb. Neurosci Res. 2015;98:28–34.
Chuang JH, Li H. Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome. PLoS Biol. 2004;2:E29.
Acknowledgements
We thank all the families for their participation in this study. This work was supported by Ongwanada Special Operating Research Fund for the genetic study of autism and related disorders and through private donations.
Author information
Authors and Affiliations
Contributions
CPS and XL designed experiment; MLH recruited participants and collected samples; AJM and SW performed next generation sequencing; CPS and SW analyzed data; CPS and ST validated variants and expression; CPS drafted manuscript; all authors edited manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sjaarda, C.P., Wood, S., McNaughton, A.J.M. et al. Exome sequencing identifies de novo splicing variant in XRCC6 in sporadic case of autism. J Hum Genet 65, 287–296 (2020). https://doi.org/10.1038/s10038-019-0707-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0707-0
This article is cited by
-
Correlated evolution of social organization and lifespan in mammals
Nature Communications (2023)
-
A Review on the Role of Genetic Mutations in the Autism Spectrum Disorder
Molecular Neurobiology (2023)