Abstract
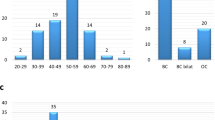
Partner and localiser of BRCA2 forms part of a macromolecular complex with BRCA1 and BRCA2, which is critical for the repair of double-strand DNA breaks by homologous DNA recombination. Germline loss-of-function variants in the PALB2 gene may confer an increased lifetime risk of breast, pancreatic, ovarian and other cancers. However, the complete spectrum of predicted pathogenic PALB2 variants associated with each tissue type of cancer remains unknown. A systematic review is performed with the aim of cataloguing predicted pathogenic PALB2 variants in breast, ovary and pancreas cancers. All catalogued predicted pathogenic variants are analysed to assess for overlap and mutational “hotspots” within gene exons. Our results showed that 911 (92.5%) cases were described in breast cancer patients, 49 (5.0%) cases were described in ovarian cancer patients, and 24 (2.4%) cases were described in pancreatic cancer patients. The top five most frequently reported predicted pathogenic PALB2 variants were c.509_510delGA, c.3113G > A, c.1592delT, c.172_175delTTGT, and c.1240C > T, accounting for 57.3% of all cases. Breast and pancreatic cancers share five variants while breast and ovarian cancers share 12 variants. Breast, ovarian and pancreatic cancers share eight common variants. Exons with the highest mutation rates were exons 2 (6.7%), 1 (6.3%) and 3 (5.8%). This systematic review provides a quantitative catalogue of predicted pathogenic PALB2 variants described in cancers. This comprehensive analysis of the PALB2 mutational spectrum represents a useful resource for clinicians overseeing PALB2-related cancer surveillance and provides a valuable resource for future PALB2-specific research.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Park JY, Zhang F, Andreassen PR. PALB2: the hub of a network of tumor suppressors involved in DNA damage responses. Biochimica Et Biophysica Acta. 2014;1846:263–75.
Zhang F, Ma JL, Wu JX, Ye L, Cai H, Xia B, et al. PALB2 Links BRCA1 and BRCA2 in the DNA-Damage Response. Curr Biol. 2009;19:524–9.
Buisson R, Dion-Cote AM, Coulombe Y, Launay H, Cai H, Stasiak AZ, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17:1247.
Casadei S, Norquist BM, Walsh T, Stray S, Mandell JB, Lee MK, et al. Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer. Cancer Res. 2011;71:2222–9.
Slater EP, Langer P, Niemczyk E, Strauch K, Butler J, Habbe N, et al. PALB2 mutations in European familial pancreatic cancer families. Clin Genet. 2010;78:490–4.
Norquist BM, Harrell MI, Brady MF, Walsh T, Lee MK, Gulsuner S, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2:482–90.
Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:18032–7.
Pearlman R, Frankel WL, Swanson B, Zhao WQ, Yilmaz A, Miller K, et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol. 2017;3:464–71.
Sabbaghian N, Kyle R, Hao A, Hogg D, Tischkowitz M. Mutation analysis of the PALB2 cancer predisposition gene in familial melanoma. Fam Cancer. 2011;10:315–7.
Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. New Engl J Med. 2016;375:443–53.
Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Sci (New Y, NY). 2009;324:217.
Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkas K, Roberts J, et al. Breast-cancer risk in families with mutations in PALB2. New Engl J Med. 2014;371:497–506.
Daly MB, Pilarski R, Berry M, Buys SS, Friedman S, Garber JE, et al. NCCN guidelines insights: genetic/familial high-risk assessment: breast and ovarian, Version 1.2019. 2018. https://www.nccn.org/professionals/physician_gls/default.aspx#detection.
O’Reilly D, Fou L, Hasler E, Hawkins J, O’Connell S, Pelone F, et al. Diagnosis and management of pancreatic cancer in adults: A summary of guidelines from the UK National Institute for Health and Care Excellence. Pancreatology. 2018;18:962–70.
Moher D, Liberati A, Tetzlaff J, Altman DG, Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097.
Corso G, Feroce I, Intra M, Toesca A, Magnoni F, Sargenti M, et al. BRCA1/2 germline missense mutations: a systematic review. Eur J Cancer Prev: Off J Eur Cancer Prev Organ (ECP). 2018;27:279–86.
Norquist BM, Harrell MI, Brady MF, Walsh T, Lee MK, Gulsuner S, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2:482–90.
Kanchi KL, Johnson KJ, Lu C, McLellan MD, Leiserson MD, Wendl MC, et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun. 2014;5:3156.
Arvai KJ, Roberts ME, Torene RI, Susswein LR, Marshall ML, Zhang Z, et al. Age-adjusted association of homologous recombination genes with ovarian cancer using clinical exomes as controls. Hereditary cancer Clin Pract. 2019;17:19.
Venkitaraman AR. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Sci (New Y, NY). 2014;343:1470–5.
Southey MC, Teo ZL, Dowty JG, Odefrey FA, Park DJ, Tischkowitz M, et al. A PALB2 mutation associated with high risk of breast cancer. Breast Cancer Res. 2010;12:1–10.
Pauty J, Couturier AM, Rodrigue A, Caron M-C, Coulombe Y, Dellaire G, et al. Cancer-causing mutations in the tumor suppressor PALB2 reveal a novel cancer mechanism using a hidden nuclear export signal in the WD40 repeat motif. Nucleic Acids Res. 2017;45:2644–57.
Erkko H, Xia B, Nikkila J, Schleutker J, Syrjakoski K, Mannermaa A, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–9.
Dansonka-Mieszkowska A, Kluska A, Moes J, Dabrowska M, Nowakowska D, Niwinska A, et al. A novel germline PALB2 deletion in Polish breast and ovarian cancer patients. BMC Med Genet. 2010;11:20.
Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci USA. 2009;106:7155–60.
Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol cancer Res: MCR. 2009;7:1110–8.
Rebbeck TR, Friebel TM, Friedman E, Hamann U, Huo D, Kwong A, et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum Mutat. 2018;39:593–620.
Hanenberg H, Andreassen P. PALB2 (partner and localizer of BRCA2). Atlas Genet Cytogenet Oncol Haematol. 2018;22.
Pauty J, Rodrigue A, Couturier A, Buisson R, Masson JY. Exploring the roles of PALB2 at the crossroads of DNA repair and cancer. Biochemical J. 2014;460:331–42.
Diamond JM, Rotter JI. Observing the founder effect in human evolution. Nature. 1987;329:105–6.
Acknowledgements
S-S Liau was funded by the Medical Research Council (MRC) and Academy of Medical Sciences Tenure-track Clinician Scientist Fellowship (Grant No: G1002543/1), Royal College of Surgeons of England Pump-priming Grant, Royal College of Surgeons of Edinburgh Small Research Grant and Pancreatic Cancer UK Research Innovation Award.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Janssen, B., Bellis, S., Koller, T. et al. A systematic review of predicted pathogenic PALB2 variants: an analysis of mutational overlap between epithelial cancers. J Hum Genet 65, 199–205 (2020). https://doi.org/10.1038/s10038-019-0680-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0680-7
This article is cited by
-
Genetic Considerations in the Locoregional Management of Breast Cancer: a Review of Current Evidence
Current Breast Cancer Reports (2023)
-
A commentary on germline mutations of multiple breast cancer-related genes are differentially associated with triple-negative breast cancers and prognostic factors
Journal of Human Genetics (2020)