Abstract
Pelizaeus-Merzbacher disease (PMD) is an X-linked recessive disorder caused by abnormalities in the gene PLP1. Most females harboring heterozygous PLP1 abnormalities are basically asymptomatic. However, as a result of abnormal patterns of X-chromosome inactivation, it is possible for some female carriers to be symptomatic. Whole-exome sequencing of a female patient with unknown spastic paraplegia was performed to obtain a molecular diagnosis. As a result, a de novo heterozygous single-nucleotide deletion in PLP1 [NM_000533.5(PLP1_v001):c.783del; p.Thr262Leufs*20] was identified. RNA sequencing was performed in a patient-derived lymphoblastoid cell line, confirming mono-allelic expression of the mutated allele and abnormal inactivation of the wild-type allele. The patient-derived lymphoblastoid cell line was then treated with VX680 or 5azadC, which resulted in restored expression of the wild-type allele. These two agents thus have the potential to reverse inappropriately-skewed inactivation of the X-chromosome.
Similar content being viewed by others
Introduction
To date, many genes responsible for hypomyelinating leukoencephalopathies (HLDs) have been identified [1]. HLD inheritance patterns depend on the affected genes. The proteolipid protein 1 gene (PLP1), located on Xq22.2, was first identified as the gene responsible for the most common HLD, Pelizaeus-Merzbacher disease (PMD; MIM #312080) [2]. As PLP1 is located on the X-chromosome, PMD is inherited as an X-linked recessive trait. Accordingly, most of the affected patients with PMD are male, while females harboring heterozygous PLP1 abnormalities typically remain asymptomatic. Therefore, when new female patients with HLD are encountered, genes other than PLP1 are suspected to be the cause. For example, the gap junction protein gamma 2 gene (GJC2) is related to an autosomal recessive trait [3], whereas the tubulin beta 4A class IVa gene (TUBB4A) is related to an autosomal dominant trait [4]. In addition to these genes, it is possible for heterozygous female carriers of PLP1 abnormalities to present clinical manifestations. This phenomenon is thought to be the result of abnormal patterns of X-chromosome inactivation (XCI).
Herein, we report a case of a Japanese girl with spastic paraplegia and delayed myelination, which was caused by a de novo heterozygous PLP1 frameshift mutation. Treatment of patient-derived lymphoblastoid cells with the chemical agents VX680 or 5azadC in vitro successfully reversed the abnormal XCI and restored expression of the wild-type allele.
Materials and methods
Materials
This study was performed in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of our institution. After receiving written informed consent, we obtained blood samples from the patient and both parents.
Molecular analysis
Genomic DNA was extracted from the patient’s peripheral blood using a standard protocol. Whole-exome sequencing was performed using trio samples, including parental samples as described previously [5]. Briefly, a SureSelect Human All Exon V6 kit (Agilent Technologies, Santa Clara, CA) and a HiSeq2500 (Illumina, San Diego, CA) with 125-bp paired-end reads were used for exon capture and sequencing, respectively. Reads were aligned to CRCh37 using the Burrows-Wheeler Aligner (http://bio-bwa.sourceforge.net/). Variants were called using the GATK Unified Genotyper and using ANNOVA (http://annover.openbioinfomatics.org/en/latest/).
To confirm RNA expression, a lymphoblastoid cell line (LCL) was established using Epstein–Barr virus transformation. Following this, total cellular RNA was extracted and harvested using a QIAGEN RNeazy Mini kit (QIAGEN, Hilden, Germany). A DNase mini kit (QIAGEN) was used to eliminate contaminating DNA, and pure cDNA was produced by reverse-transcription (RT) using a superscript VILO cDNA Synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA). The PLP1 cDNA was amplified using nested RT-PCR. The first and second rounds of PCR were performed using KOD-FX (Toyobo, Osaka, Japan) and TAKARA EX-Taq (Takara Bio Inc., Kusatsu, Japan), respectively. In the first PCR step, we used the following primers: PLP1 nested-F1: 5′-CAAGGTTTGTGGCTCCAACCTTCTG-3′ and PLP1 nested-R1: 5′-AGCATTGTAGGCTGTGTGGTTAGAG-3′. In the second PCR step, a different set of primers was used (PLP1 nested-F2: 5′-CCATCTGCAAAACAGCTGAG-3′ and PLP1 nested-R2: 5′-GCCTCGCTATTAGAGAAAGG-3′).
XCI status was analyzed through methylation-specific PCR of the human androgen receptor gene (HUMARA), as described elsewhere [6, 7].
Manipulation of XCI status
We performed in vitro drug treatment of the LCL derived from the patient to determine whether the XCI status could be treated in accordance with previously reported methods [8]. Two compounds were used: VX680 (an Aurora kinase inhibitor) and 5azadC (a methyltransferase inhibitor). Both chemicals were dissolved in dimethyl sulfoxide (DMSO) to create 5 mM stocks. The LCL was seeded at 60,000 cells per well in six-well plates on day 2. Drug treatment occurred 48 h after seeding, on day 0. In addition to the DMSO control, we treated the patient’s LCL with either VX680 or 5azadC. The drug concentrations used were 2.5, 5, and 10 μM. Total cellular RNA was then harvested on day 3. Subsequently, RNA sequencing was performed using RT-PCR amplicons as described above. These experiments were performed on three independent replicates. As the expression levels could not be measured with Sanger sequencing, the results were qualitatively evaluated to determine whether they showed mono-allelic or bi-allelic expression levels.
Results
Patient reports
A 7-year-old girl is the second child of healthy, non-consanguineous, Japanese parents. Her elder sister is healthy. After an uneventful pregnancy, the patient was born at 38 weeks of gestation with a birth weight of 2908 g. She was able to hold her head up at 5 months, roll over on her other side at 6 months, and walk with support at 12 months of age. As she could not walk unsupported by the age of 20 months, her parents took her to a hospital for a consultation. At that time, mild spasticity was observed. Brain magnetic resonance imaging (MRI) first revealed the patient’s delayed neuronal myelination (Fig. 1a); mildly delayed neuronal myelination was also observed at 3 years of age (Fig. 1b). From these findings, she was diagnosed with spastic paraplegia associated with delayed myelination. At 5 years of age, her intelligence quotient was evaluated to be 82. However, she was suspected of having attention-deficit hyperactivity disorder owing to observed distraction and disconnection. Behavioral abnormalities, such as aggressive behavior and self-injurious activity were not observed.
At present, her height is 114.4 cm (−1.1 SD), and her weight is 21.4 kg (−0.4 SD). She still shows clinical signs of spastic paraplegia, but no symptoms of nystagmus or cerebellar ataxia are apparent. There are no dysmorphic features and no visceral abnormalities. She has never had episodes of epilepsy. She has a normal female karyotype of 46,XX. Duplication of the peripheral myelin protein 22 gene (PMP22) was not observed by fluorescence in situ hybridization analysis. Findings from brainstem auditory evoked potentials and spinal MRI were unavailable.
Experimental analysis
Whole-exome sequencing using trio samples revealed a de novo single-nucleotide deletion: NM_000533.5(PLP1_v001):c.783del; p.Thr262Leufs*20. This finding was confirmed by Sanger sequencing (Fig. 2a). The identified deletion was located on exon 7, which is the last exon in PLP1. As most females with PLP1 mutations are asymptomatic carriers, the patient’s XCI status was analyzed using DNA extracted from the patient’s peripheral blood. The patient showed only a single peak of the PCR amplicon for HUMARA (data not shown). This indicates a homozygous pattern for this variant and, as a result, we could not identify, which allele was activated. Thus, HUMARA was an inappropriate analytical method for assessing XCI in this patient.
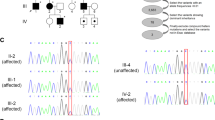
Results of the molecular analyses. a Electropherograms of Sanger sequencing for PLP1 exon 7. Genomic DNA derived from the patient and her parents were used for sequencing. Only the patient showed a single-nucleotide deletion, indicating that this is a de novo occurrence. b Sanger sequencing of RT-PCR products only shows the presence of the mutated allele. c RNA sequencing after chemical treatment demonstrates the successful reactivation of silenced wild-type alleles (asterisks). I; control, II; 5-aza-dC 2.5 μM, III; VX680 2.5 μM, and IV; 5-aza-dC 5 μM
For the reason stated above, RNA sequencing of PLP1 in the patient’s LCL was performed. As the expression of PLP1 in LCLs is cryptic [9] and PLP1 mRNA is difficult to amplify in a single round of PCR, a nested-PCR method was used instead [10]. Sanger sequencing of the amplified PLP1 showed only the sequence derived from the mutated allele (Fig. 2b). This indicated that there was an unfavorable skewed inactivation of the wild-type allele, leading to clinical manifestations in this female patient. Generally, skewed inactivation is indicated when one of two alleles is predominantly inactivated by >80%.
Next, an in vitro experiment was performed to attempt to reverse the XCI by treating the patient-derived LCL with VX680 or 5azadC. After culturing, the number of live cells was slightly reduced by the addition of VX680 or 5-aza-dC. This may indicate that these compounds have cytotoxic effects (Table 1). After harvesting the cells, bi-allelic PLP1 mRNA expression of both wild-type and mutated alleles was confirmed in multiple replicates (Fig. 2c), indicating successful reactivation of the wild-type allele.
Discussion
In this study, whole-exome sequencing was performed in a female patient with spastic paraplegia associated with delayed myelination, and a de novo PLP1 mutation was identified. However, it was uncertain whether or not the identified heterozygous PLP1 mutation was actually related to the patient’s phenotype, because most females with heterozygous PLP1 mutations are clinically recognized to be asymptomatic, healthy carriers. However, in rare cases, female carriers may develop clinical symptoms, with great variability.
Until now, at least 19 female patients have been reported as symptomatic carriers of PLP1 abnormalities [11,12,13,14,15,16,17,18,19,20,21] (Table 2). Some patients showed only spasticity, but other patients showed full manifestations of PMD. Very rare cases of late-onset spastic paraplegia and adult-onset progressive leukodystrophy with dementia have been also reported [22]. Such clinical variability is considered to be a result of variable XCI patterns, which can affect the phenotypic severity, ranging from severe to transiently symptomatic or asymptomatic [13].
Intra-familial clinical variabilities have also been noted. In 1995, Hodes et al. [11] reported three carrier females amongst three generations of a family. Among them, only one female child was affected by motor developmental delay and hypomyelination, while her mother and grandmother (also carriers) showed no symptoms related to PLP1 abnormalities. This intra-familial clinical variability is also caused by variable XCI patterns in individuals.
Previously, there were six reports in which carrier females were analyzed for their XCI status (Table 2). As shown by Woodward et al. [12], nine asymptomatic carrier females showed skewed XCI patterns in favor of the wild-type gene. This indicates that asymptomatic carrier females escape clinical onset through near-complete silencing of the alleles containing PLP1 abnormalities. On the other hand, six affected females possessed random (not skewed) XCI patterns. In these cases, affected alleles are randomly activated in cells and patients are unable to escape clinical manifestations. However, XCI patterns do not completely correlate with clinical symptoms; as reported by Woodward et al. [12], three asymptomatic carrier females had random (not skewed) XCI patterns. Furthermore, as shown by Carvalho et al. [16], XCI patterns are variable among different tissues (XCI patterns were different in blood and buccal cells derived from the same individuals). Thus, there is no consistency in XCI patterns and an examination of XCI may be an unsuitable diagnostic tool for female carriers of PMD [12].
In this study, the mRNA expression of PLP1 in a patient-derived LCL was analyzed, revealing that only the mutated allele was expressed. This suggests that the wild-type PLP1 allele is completely inactivated, and only the mutated allele is activated. Thus, skewed XCI in this patient was considered to be the reason this patient is symptomatic, and the final diagnosis of the patient as a symptomatic carrier of a PLP1 abnormality was made.
Although the single-nucleotide deletion identified in this study has never been reported previously, it is considered to be disease-causing as it is predicted to lead to a frameshift. Generally, mRNAs containing premature termination codons are degraded by nonsense-mediated RNA decay (NMD), which leads to a loss-of-function of the protein. However, mRNAs with premature termination codons near the stop codon can escape NMD [23]. The deletion identified in this study is located in the last exon and near the stop codon, causing a 60-bp (20 amino-acid) elongation from the site of the single-nucleotide deletion and creating a new termination codon. Thus, the total length of the abnormally elongated PLP1 mRNA in this patient is 846-bp (estimated to be 282 amino acids), compared to the normal PLP1 mRNA length of 831-bp (277 amino acids). This abnormal PLP1 mRNA may escape NMD and result in the expression of an abnormal PLP1 protein, which could provide a gain-of-function rather than a loss-of-function of PLP1 in cells.
It is well known that the clinical severity of PMD in patients is related to the different types of PLP1 abnormalities [2, 22]. Connatal PMD (the most severe type) is caused by the cytotoxic effects of misfolded PLP1 in oligodendrocytes, which is mostly due to missense mutations, indicating a gain-of-function mechanism. Classical PMD (the milder and most common type) is generally caused by PLP1 duplications resulting in excess protein, indicating a gain-of-function mechanism similar to that found in the connatal type. PLP1 null mutations, including large deletions and nonsense mutations, are related to spastic paraplegia (considered to be a milder form of PMD). However, all of these classifications are applied only in cases of male PMD patients. In cases of female carriers with clinical manifestations, clinical severity depends instead on XCI patterns. Thus, in the current female patient, we cannot determine if the mechanism of disease is a gain-of-function or loss-of-function.
Previously, several studies have been performed to treat abnormal XCI patterns in patients. For this purpose, several epigenetic activators have been tested for their ability to reactivate the expression of genes that have been silenced by XCI [8, 24,25,26]. Confirming previous studies’ findings, we were able to reactivate the silenced wild-type allele in cultures of the patient’s LCL through treatment with VX680 or 5azadC. As VX680 and 5azadC have cytotoxic effects, these compounds cannot be clinically used in patients. However, this study suggests that in the future there is potential for other small molecules to be developed for the treatment of disorders related to abnormal XCI.
References
Charzewska A, Wierzba J, Izycka-Swieszewska E, Bekiesinska-Figatowska M, Jurek M, Gintowt A, et al. Hypomyelinating leukodystrophies-a molecular insight into the white matter pathology. Clin Genet. 2016;90:293–304.
Yamamoto T, Shimojima K. Pelizaeus-Merzbacher disease as a chromosomal disorder. Congenit Anom (Kyoto). 2013;53:3–8.
Shimojima K, Tanaka R, Shimada S, Sangu N, Nakayama J, Iwasaki N, et al. A novel homozygous mutation of GJC2 derived from maternal uniparental disomy in a female patient with Pelizaeus-Merzbacher-like disease. J Neurol Sci. 2013;330:123–6.
Shimojima K, Okumura A, Ikeno M, Nishimura A, Saito A, Saitsu H, et al. A de novo TUBB4A mutation in a patient with hypomyelination mimicking Pelizaeus-Merzbacher disease. Brain Dev. 2015;37:281–5.
Sasaki H, Yanagi K, Ugi S, Kobayashi K, Ohkubo K, Tajiri Y, et al. Definitive diagnosis of mandibular hypoplasia, deafness, progeroid features and lipodystrophy (MDPL) syndrome caused by a recurrent de novo mutation in the POLD1 gene. Endocr J. 2018;65:227–38.
Bertelsen B, Tumer Z, Ravn K. Three new loci for determining x chromosome inactivation patterns. J Mol Diagn. 2011;13:537–40.
Shimada S, Okamoto N, Ito M, Arai Y, Momosaki K, Togawa M, et al. MECP2 duplication syndrome in both genders. Brain Dev. 2013;35:411–9.
Yu D, Sakurai F, Corey DR. Clonal Rett Syndrome cell lines to test compounds for activation of wild-type MeCP2 expression. Bioorg Med Chem Lett. 2011;21:5202–5.
Shimojima K, Inoue T, Imai Y, Arai Y, Komoike Y, Sugawara M, et al. Reduced PLP1 expression in induced pluripotent stem cells derived from a Pelizaeus-Merzbacher disease patient with a partial PLP1 duplication. J Hum Genet. 2012;57:580–6.
Omata T, Nagai J, Shimbo H, Koizume S, Miyagi Y, Kurosawa K, et al. A splicing mutation of proteolipid protein 1 in Pelizaeus-Merzbacher disease. Brain Dev. 2016;38:581–4.
Hodes ME, DeMyer WE, Pratt VM, Edwards MK, Dlouhy SR. Girl with signs of Pelizaeus-Merzbacher disease heterozygous for a mutation in exon 2 of the proteolipid protein gene. Am J Med Genet. 1995;55:397–401.
Woodward K, Kirtland K, Dlouhy S, Raskind W, Bird T, Malcolm S, et al. X inactivation phenotype in carriers of Pelizaeus-Merzbacher disease: skewed in carriers of a duplication and random in carriers of point mutations. Eur J Hum Genet. 2000;8:449–54.
Inoue K, Tanaka H, Scaglia F, Araki A, Shaffer LG, Lupski JR. Compensating for central nervous system dysmyelination: females with a proteolipid protein gene duplication and sustained clinical improvement. Ann Neurol. 2001;50:747–54.
Fattal-Valevski A, DiMaio MS, Hisama FM, Hobson GM, Davis-Williams A, Garbern JY, et al. Variable expression of a novel PLP1 mutation in members of a family with Pelizaeus-Merzbacher disease. J Child Neurol. 2009;24:618–24.
Yiu EM, Farrell SA, Soman T. Classic Pelizaeus-Merzbacher disease in a girl with an unbalanced chromosomal translocation and functional duplication of PLP1. Mov Disord. 2009;24:2171–2.
Carvalho CM, Bartnik M, Pehlivan D, Fang P, Shen J, Lupski JR. Evidence for disease penetrance relating to CNV size: Pelizaeus-Merzbacher disease and manifesting carriers with a familial 11 Mb duplication at Xq22. Clin Genet. 2012;81:532–41.
Fonseca AC, Bonaldi A, Costa SS, Freitas MR, Kok F, Vianna-Morgante AM. PLP1 duplication at the breakpoint regions of an apparently balanced t(X;22) translocation causes Pelizaeus-Merzbacher disease in a girl. Clin Genet. 2013;83:169–74.
Matsufuji M, Osaka H, Gotoh L, Shimbo H, Takashima S, Inoue K. Partial PLP1 deletion causing X-linked dominant spastic paraplegia type 2. Pedia Neurol. 2013;49:477–81.
Lassuthova P, Zaliova M, Inoue K, Haberlova J, Sixtova K, Sakmaryova I, et al. Three new PLP1 splicing mutations demonstrate pathogenic and phenotypic diversity of Pelizaeus-Merzbacher disease. J Child Neurol. 2014;29:924–31.
Brender T, Wallerstein D, Sum J, Wallerstein R. Unusual presentation of pelizaeus-merzbacher disease: female patient with deletion of the proteolipid protein 1 gene. Case Rep Genet. 2015;2015:453105.
Masliah-Planchon J, Dupont C, Vartzelis G, Trimouille A, Eymard-Pierre E, Gay-Bellile M, et al. Insertion of an extra copy of Xq22.2 into 1p36 results in functional duplication of the PLP1 gene in a girl with classical Pelizaeus-Merzbacher disease. BMC Med Genet. 2015;16:77.
Inoue K. PLP1-related inherited dysmyelinating disorders: Pelizaeus-Merzbacher disease and spastic paraplegia type 2. Neurogenetics. 2005;6:1–16.
Inoue K, Khajavi M, Ohyama T, Hirabayashi S, Wilson J, Reggin JD, et al. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet. 2004;36:361–9.
Bhatnagar S, Zhu X, Ou J, Lin L, Chamberlain L, Zhu LJ, et al. Genetic and pharmacological reactivation of the mammalian inactive X chromosome. Proc Natl Acad Sci USA. 2014;111:12591–8.
Minkovsky A, Sahakyan A, Bonora G, Damoiseaux R, Dimitrova E, Rubbi L, et al. A high-throughput screen of inactive X chromosome reactivation identifies the enhancement of DNA demethylation by 5-aza-2’-dC upon inhibition of ribonucleotide reductase. Epigenetics Chromatin. 2015;8:42.
Lessing D, Dial TO, Wei C, Payer B, Carrette LL, Kesner B, et al. A high-throughput small molecule screen identifies synergism between DNA methylation and Aurora kinase pathways for X reactivation. Proc Natl Acad Sci USA. 2016;113:14366–71.
Acknowledgements
We would like to express our gratitude to the patient and her family for their cooperation. This work was supported by a Grant-in-Aid for Young Scientists (B) (17K18133) and a Restart Postdoctoral Fellowship (17J40108) from the Japan Society for the Promotion of Science (JSPS) for KY. This study was also funded by the Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development (AMED 18ek0109270, TY, KI) and JSPS KAKENHI JP18K07803 (TY). We are also thankful for the support from the Initiative on Rare and Undiagnosed Diseases (IRUD) via the Japan Agency for Medical Research and Development (AMED).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yamamoto-Shimojima, K., Imaizumi, T., Aoki, Y. et al. Elucidation of the pathogenic mechanism and potential treatment strategy for a female patient with spastic paraplegia derived from a single-nucleotide deletion in PLP1. J Hum Genet 64, 665–671 (2019). https://doi.org/10.1038/s10038-019-0600-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0600-x
This article is cited by
-
Novel BCL11B truncation variant in a patient with developmental delay, distinctive features, and early craniosynostosis
Human Genome Variation (2022)
-
Whole-exome analysis of 177 pediatric patients with undiagnosed diseases
Scientific Reports (2022)
-
Deep intronic deletion in intron 3 of PLP1 is associated with a severe phenotype of Pelizaeus-Merzbacher disease
Human Genome Variation (2021)
-
A recurrent de novo ZSWIM6 variant in a Japanese patient with severe neurodevelopmental delay and frequent vomiting
Human Genome Variation (2021)
-
Novel LAMA2 variants identified in a patient with white matter abnormalities
Human Genome Variation (2020)