Abstract
Insertion and deletion markers (InDels) have gained considerable attentions in population genetics and forensic research. In this study, we investigated genetic distributions of 30 InDels in Gansu Yugur and Guizhou Miao groups and evaluated their forensic application values. Genetic relationship analyses between Gansu Yugur, Guizhou Miao groups and other published populations were conducted based on these 30 InDels. Power of discrimination and power of exclusion in trio and duo cases of 30 InDels ranged from 0.3528 to 0.6247, 0.0937 to 0.1873, and 0.0219 to 0.1247 in Gansu Yugur group; and they ranged from 0.2579 to 0.6247, 0.0671 to 0.1874, and 0.0105 to 0.1247 in Guizhou Miao group. Obtained cumulative power of discrimination values indicated these InDels could be used for forensic individual identifications in both ethnic groups. Principal component analysis and phylogenetic reconstruction revealed that Gansu Yugur and Guizhou Miao groups had close affinities with their neighboring populations. Genetic structure analyses among these populations also indicated that studied Gansu Yugur and Guizhou Miao groups showed similar genetic structure with their neighboring populations. Further analyses of Y-STR, mtDNA, and ancestry informative markers should be conducted to better understand genetic backgrounds of Gansu Yugur and Guizhou Miao groups in the future.
Similar content being viewed by others
Introduction
Short tandem repeats (STRs) are commonly used genetic markers in forensic investigation since they are highly polymorphic [1]. However, STR profiles of some samples like highly degraded or dated biological materials may not be obtained. The presence of stutter peaks of STR profiles may mislead the interpretation of mixed samples [2]. Besides, high mutation rate of STRs may induce adverse effects in paternity testing [3]. Contrary to STRs, insertion and deletion (InDel) polymorphisms possess some advantages including low mutation rate [4] and small amplicons [5], which has been paid considerable attentions by forensic geneticists during the recent years. For examples, some researchers constructed InDels panels for ancestral origin estimation [6,7,8]; multiplex InDels systems for forensic individual identification have been presented [9, 10].
As the first commercial InDels kit (Investigator DIPplex Kit) having been released, an army of population data regarding these InDel markers were reported [11,12,13,14,15,16,17,18,19,20,21,22,23]. Previous validation analyses of the kit have shown that the kit could type low amount DNA samples (62 pg) and degraded casework samples; their results stated that the assay could be viewed as a useful tool for human individual identification [2, 24]. As genetic variations of the same genetic markers in various populations exist, it is vital for us to evaluate their genetic distributions in the studied population before these markers being used for forensic applications [25].
Yugur group is one of 55 ethnic groups in China, which mainly distributes in Gansu province. The language of Yugur people belongs to the branch of the Altaic language family. Although Yugur group has relatively small population size, it has a long history. For ethnic origins of Yugur group, frequent interactions between Yugur group and its neighboring populations exert certain impacts on the formation of Yugur group [26]. Accordingly, research on genetic background of Yugur group should be performed, which will benefit for revealing its ethnic origin. Genetic distributions of autosomal STRs [27], X-STRs [25], and HLA alleles [28] in Yugur group have been conducted, but no report on InDel markers is published. Likewise, Miao group is also an old ethnic group in China, which mainly dwells in south China regions like Guangxi Zhuang Autonomous region and Yunnan, Guizhou, and Sichuan provinces. Miao people cultivated millet and buckwheat with the method of the slash-and-burn before modernization of farming methods have been putting into practice. Their religion beliefs include nature and ancestor worship and Christianity. The language of Miao people is part of the Hmong-Yao branch of Sino-Tibetan language family (http://www.paulnoll.com/China/Minorities/min-Miao.html). Although research on InDels in Miao group has been conducted [12], further analysis in Miao samples from different regions may contribute to revealing its genetic background better.
To enrich genetic data of Gansu Yugur and Guizhou Miao groups, genetic diversities of 30 InDels in both populations were assessed on the basis of Investigator DIPplex Kit (Qiagen, Hilden, Germany). Forensic efficiencies of these markers were evaluated. Moreover, genetic relationship analyses between the studied Gansu Yugur, Guizhou Miao groups, and other published populations [11,12,13,14,15,16,17,18,19,20,21,22,23, 29,30,31] were performed.
Materials and methods
Sample collections and DNA extraction
The study was in conformity with the human and ethical research principles of Xi’an Jiaotong University Health Science Center, and was warranted by the ethics committee of Xi’an Jiaotong University Health Science Center. Prior to sample collections, written informed consent of each participant was obtained. Blood-stain samples of 139 Yugur individuals living in Sunan Yugur Autonomous County, Gansu province and 274 Miao individuals residing in Guizhou province were gathered. DNA samples of the studied individuals were extracted according to the Chelex-100 method [32]. Supplementary Fig. 1 showed geographical distributions of Gansu Yugur, Guizhou Miao, and other 22 reference populations.
Multiplex PCR amplification and InDel genotyping
Referring to the specification provided by the kit, multiplex PCR amplification of 30 InDels and the Amelogenin gene was conducted. Next, the cocktail including 1 µL PCR product and the mixture of 12 µL Hi-Di formamide and 0.5 µL DNA Size Standard 550 (BTO) was initially denatured at 95 °C for 3 min, then immediately chilled at 4 °C for 5 min. Allelic assignments of 30 InDels and the Amelogenin gene were conducted on the ABI 3500xL Genetic Analyzer instrument (Applied Biosystems, USA) according to the parameters recommended by the kit. Finally, alleles were determined on GeneMapper ID software v3.2 (Applied Biosystems, USA) by comparing with the allele ladder of the kit. During the experimental process, control DNA 9948 and nuclear-free water were used as positive and negative controls, respectively. The genotyping results of 30 InDels and the Amelogenin gene in the studied Gansu Yugur and Guizhou Miao groups were given in Supplementary Table 1.
Statistical analysis
STR_Genotype tool [33] was used to calculate allele frequencies and forensic relevant parameters of 30 InDels. Hardy–Weinberg equilibrium (HWE) analyses of 30 InDels were performed by Arlequin software v3.5 [34]. Linkage disequilibrium (LD) analyses (correlation coefficient, r2) of pairwise InDels were estimated by SHEsis program [35]. A heatmap of insertion allele frequencies for 30 InDels in the studied Gansu Yugur, Guizhou Miao groups and other compared populations was plotted by pheatmap program in R software v3.3 [36]. Average heterozygosity values and genetic distances (DA) of Gansu Yugur, Guizhou Miao groups and other reference populations were calculated by DISPAN program (https://www.winsite.com/Multimedia/3D-Modeling-CAD/DISPAN/) based on allele frequencies of 30 InDels. To explore genetic affinities among Gansu Yugur, Guizhou Miao groups and other reference populations, principal component analysis (PCA) was performed by XLSTAT program (https://www.xlstat.com/en/). Based on DA values among these populations, a phylogenetic tree was constructed by MEGA software v6.0 [37] based on the Neighbor-Joining method. Assuming admixture, LOCPRIOR and correlated allele frequencies model, genetic structure analyses of 24 populations were run five replicates for K = 2–5 with the 10,000 burnins and 10,000 MCMC by STRUCTURE software v2.3 [38]. Estimated individuals’ genetic components were displayed in the bar plot by distruct program [39].
Results and discussion
HWE and LD analyses for 30 InDels
The P-values for HWE tests of 30 InDels in Gansu Yugur and Guizhou Miao groups were given in Supplementary Tables 2–3. Although the P-values of rs1610905 locus in Gansu Yugur group and rs6481 locus in Guizhou Miao group were less than 0.05, P-values of all loci were not statistically significant after applying Bonferroni correction (the significant level = 0.05/30 = 0.0017), implying these loci conformed to HWE in Gansu Yugur and Guizhou Miao groups. Therefore, these 30 InDels could be employed for the further analysis.
LD indicated that alleles at different loci were in the state of non-random associations. In forensic research, LD analyses of pairwise loci should be evaluated prior to product law being employed. Correlation coefficient (r2) could be utilized to evaluate the level of LD between pairwise loci: pairwise loci will have strong correlations when the r2 value between them is more than 0.8 [40]. The r2 values of pairwise 30 InDels in Gansu Yugur and Guizhou Miao groups were displayed in Supplementary Figs. 2 and 3. These r2 values were magnified 100 times so that we could display them easily. Results showed that r2 values of all pairwise loci were less than 0.1, indicating relatively weak correlations among these pairwise loci. Consequently, these loci could be viewed as independent loci from each other in Gansu Yugur and Guizhou Miao groups.
Allele frequencies and forensic parameters of 30 InDels in Gansu Yugur and Guizhou Miao groups
Allele frequencies of 30 InDels were shown in Supplementary Fig. 4. For biallelic markers, the markers can be used as valuable loci for forensic individual identification as long as their minor allele frequencies are more than 0.2 [10]. In Gansu Yugur group, the minor allele frequencies of five InDels (rs17878444, rs1610935, rs2308163, rs1305047, and rs16438) were less than 0.2, demonstrating these loci were less valuable in Gansu Yugur group (Supplementary Fig. 4a). For Guizhou Miao group, rs17878444, rs1610935, rs2308163, rs1305047, rs16438, and rs8178524 loci showed less valuable (Supplementary Fig. 4b). Therefore, more loci which are suitable for forensic individual identifications in Chinese populations should be selected in the future.
Forensic relevant parameters including observed heterozygosity (Ho), expected heterozygosity (He), polymorphism information content (PIC), power of discrimination (PD), power of exclusion in trio (PEtrio), and power of exclusion in duo (PEduo) were shown in Fig. 1. Detailed values of these parameters were given in Supplementary Tables 2 and 3. For Gansu Yugur group, the highest He, PIC, PD, PEtrio, and PEduo values were observed at rs1610905 locus with the values of 0.4994, 0.3747, 0.6247, 0.1873, and 0.1247, respectively; whereas the lowest He, PIC, PD, PEtrio, and PEduo values were at rs16438 locus (Fig. 1a). For Guizhou Miao group (Fig. 1b), the highest Ho, He, PIC, PD, PEtrio, and PEduo values were observed at rs2307652 locus, and the lowest values were at rs16438 locus. Obtained CPD, CPE in trio cases and CPE in duo cases for 30 InDels in Gansu Yugur group were 0.99999999999521, 0.9961, and 0.9549, respectively. And these values in Guizhou Miao group were 0.999999999961336, 0.9940, and 0.9346, respectively. PIC indicates that one locus can provide genetic information content: the locus with PIC value more than 0.50 is highly informative; the locus with PIC value ranging from 0.25 to 0.50 is reasonably informative; the locus with PIC value less than 0.25 is slight informative [41]. In this study, three InDel loci (rs16438, rs1305047, and rs2308163) with PIC values less than 0.25 were observed in Gansu Yugur group; and six loci, i.e. rs16438, rs2308163, rs17878444, rs1305047, rs1610935, and rs8178524 in Guizhou Miao group. Not surprisingly, these 30 InDel loci showed relatively low PIC values when comparing with the results of autosomal STRs in Yugur [27] and Miao groups [42], which resulted from inherently limited discriminations of biallelic markers. Even so, obtained CPD values revealed these loci met the requirement of forensic individual investigations in Gansu Yugur and Guizhou Miao groups. However, CPE values of 30 InDels reflected that these loci might be employed as the supplementary system for paternity tests in Gansu Yugur and Guizhou Miao groups.
Forensic relevant parameters of 30 InDels in the Gansu Yugur (a) and Guizhou Miao (b) groups. The abbreviations including Ho, He, PIC, PD, PEtrio, and PEduo denote observed heterozygosity, expected heterozygosity, polymorphism information content, power of discrimination, power of exclusion in trio case, power of exclusion in duo case, respectively. Red and blue stars denote the highest and smallest values, respectively. (color figure online)
Frequency distribution comparisons of 30 InDels in Gansu Yugur, Guizhou Miao groups, and other reference populations
As shown in Fig. 2, a heatmap of insertion allele frequencies of 30 InDels was constructed to analyze genetic variations of these loci in different populations. Color contrasts of pairwise populations demonstrated their genetic variations: more obvious color contrast of pairwise populations was, the larger their genetic differentiations were. The results revealed most loci showed similar frequency distributions in these populations. However, rs1305047, rs16438, rs2308163, rs1610935, and rs17879936 loci were found to show higher or lower allele frequencies in European populations including Dane, Hungarian, Basque, and Central Spanish when compared with other populations, indicating that these loci might be good for discriminating these European populations from the other populations. Genetic distribution research of these 30 InDels also observed that rs1305047 and rs16438 loci showed relatively high genetic differentiations among different populations, and they proposed that these loci could be employed to bio-geographical origin analysis [24]. Therefore, rs1305047 and rs16438 loci can be considered ancestry informative loci for differentiating these populations.
Genetic heterozygosity analyses of 24 populations based on 30 InDels
Heterozygosity could be used to evaluate genetic variation of certain population: the higher heterozygosity of the population was, the larger genetic variation in population was [27]. Average heterozygosity comparisons of 24 populations were conducted based on allele frequencies of 30 InDel loci (Table 1). Results showed that average heterozygosity values of these populations ranged from 0.3867 in Guangxi Dong group to 0.4910 in Hungarian group. High heterozygosity values were observed in Basque, Dane, Central Spanish, and Hungarian groups, whereas lower heterozygosity values were observed in most Chinese populations, implying that these 30 InDels showed lower polymorphisms in Chinese populations in comparison with European populations. Moreover, heterozygosity value in Gansu Yugur group was 0.4356, indicating relatively high heterozygosity in Gansu Yugur group. However, heterozygosity value in Guizhou Miao (0.3987) reflected low genetic heterozygosity, which was similar with the previous result (0.3976) in Miao group [12]. Lifestyle of dwelling tightly in community might lead to low heterozygosity of Miao group [43].
Genetic relationship explorations among Gansu Yugur, Guizhou Miao groups, and other reference populations
Based on allele frequencies of 30 InDels, PCA plot among Gansu Yugur, Guizhou Miao groups and other reference populations was conducted (Fig. 3). A total of 79.44% of the variance can be explained by the first two principal components. Different colors in Fig. 3 denote different language families. Results showed that some populations with the same language families did not cluster closely, like Hubei Tujia, Yunnan Yi, and Tibetan groups. Geographical isolation may contribute to genetic differentiations of these populations. Moreover, we found that three Eurasian populations (Xinjiang Kazak, Xinjiang Uygur, and Xinjiang Kyrgyz) located in the lower right part; most southern China populations including Guizhou Miao, Guangxi Dong, Yunnan Yi, Guangxi Zhuang, and Guangxi Miao positioned in the upper left corner; northwest China populations including Gansu Yugur group situated in the lower left part; four European populations located in the upper right part. In short, populations from different regions exhibited certain genetic variations, which lead to their different distribution patterns in the PCA plot.
To further analyze genetic relationships of these populations, a phylogenetic tree was constructed based on pairwise DA values (Fig. 4). Results indicated that these populations could be classified into two apparent branches: one included four European populations and Xinjiang Uygur group; the other comprised other Chinese populations, of which south to north cline was observed. The studied Gansu Yugur group firstly located in one sub-branch with Tibetan populations from different regions, and then with Qinghai Salar, Gansu Dongxiang, and other compared populations, implying Gansu Yugur group had close genetic affinities with Tibetan, Salar, and Dongxiang groups. The other studied Guizhou Miao group clustered with Guangxi Dong, Guangxi Miao, Yunnan Yi, and Guangxi Zhuang groups, revealing close genetic relationships between the studied Guizhou Miao group and its neighboring populations.
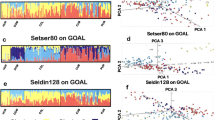
To assess ancestral components of studied Gansu Yugur and Guizhou Miao groups, genetic structure analyses of these 24 populations was conducted at K = 2–5 based on raw data of 30 InDel loci (Fig. 5). At K = 2, four European populations including Basque, Central Spanish, Dane, and Hungarian displayed orange components; Han populations from different regions, Hubei Tujia, Yunnan Yi, Guangxi Dong, Guangxi Zhuang, Guangxi Miao, and Guizhou Miao showed blue components; Xinjiang Uygur, Xinjiang Kazak, Xinjiang Kyrgyz, Tibet Tibetan, Qinghai Tibetan, Qinghai Salar, Gansu Dongxiang, and Gansu Yugur groups demonstrated the admixed components. As the increase of K values, populations from different sampling locations showed different genetic component distributions, especially for Chinese populations. For the studied Gansu Yugur group, we found it showed similar ancestral proportions with Gansu Dongxiang populations at K = 2–5. Besides, similar genetic structure among the studied Guizhou Miao, Guangxi Miao, Guangxi Zhuang, and Guangxi Dong populations were observed. Based on the present results, we found that the studied Gansu Yugur and Guizhou Miao groups showed similar ancestral components with their neighboring populations, respectively.
For ethnic origin of Yugur group, most scholars stated that ancient Huihe people living around Erhui River might be ancestors of Yugur group. After Huihe Khanate fell apart during the Tang dynasty, some survivors begun their western migration and settled in regions around Qilian mountain and Hexi corridor. As Hexi corridor being conquered by Mongolian in the 13th century, Huihe individuals living here were ruled by Mongolians. Both ethnic groups (Huihe and Mongolian group) got along with each other swimmingly and gradually formed one new group Yugur. Furthermore, the Tibetan culture and Yugur group’s surrounding ethnic groups like Tu nationality also contributed to the formation of today Yugur group [26, 44]. By assessing genetic distributions of six X-STR loci in Yugur group, Chen et al. found that Yugur group located in one branch with Tibetan, Xi’an Han and Mongolian groups in the phylogenetic tree and they suggested Yugur group was closely related with Tibetan, Mongolian, and Han populations [25]. Research on HLA allele and haplotype distributions demonstrated that Yugur group had close genetic relationships with northern China populations like Tu, Hui, Beijing Han, and Tibetan groups [28]. In this study, based on 30 InDels, close genetic relationships between Gansu Yugur group and Tibetan, Gansu Dongxiang groups were observed, which might attribute to gene and culture interactions of these populations in history. For the other studied Guizhou Miao group, close genetic relationships among the studied Guizhou Miao group and Guangxi Dong, Guangxi Miao and Guangxi Zhuang groups were observed. Previous research on autosomal STRs in Miao group also found similar results: Miao group had relatively close genetic affinities with its neighboring populations like Guangxi Yao [45], Guangxi Dong, and Guangxi Bouyei populations [46]. Further analyses of Y-STRs, mtDNA, and ancestry informative markers in Gansu Yugur and Guizhou Miao groups should be conducted, which might be beneficial for parsing their ethnic histories and population migration process.
In conclusion, we presented genetic data of 30 InDel loci in Gansu Yugur and Guizhou Miao groups, which laid the foundation for forensic and population genetic studies in the future. Obtained CPD values supported that these InDels could be used for forensic personal identification in Gansu Yugur and Guizhou Miao groups.
References
Edwards A, Civitello A, Hammond HA, Caskey CT. DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am J Hum Genet. 1991;49:746–56.
LaRue BL, Ge J, King JL, Budowle B. A validation study of the Qiagen Investigator DIPplex(R) kit; an INDEL-based assay for human identification. Int J Leg Med. 2012;126:533–40.
Brinkmann B, Klintschar M, Neuhuber F, Huhne J, Rolf B. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am J Hum Genet. 1998;62:1408–15.
Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304.
Romanini C, Catelli ML, Borosky A, Pereira R, Romero M, Salado Puerto M, et al. Typing short amplicon binary polymorphisms: supplementary SNP and Indel genetic information in the analysis of highly degraded skeletal remains. Forensic Sci Int Genet. 2012;6:469–76.
Zaumsegel D, Rothschild MA, Schneider PM. A 21 marker insertion deletion polymorphism panel to study biogeographic ancestry. Forensic Sci Int Genet. 2013;7:305–12.
Santos NPC, Ribeiro-Rodrigues EM, Ribeiro-dos-Santos AKC, Pereira R, Gusmao L, Amorim A, et al. Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat. 2010;31:184–90.
Lan Q, Shen C, Jin X, Guo Y, Xie T, Chen C, et al. Distinguishing three distinct biogeographic regions with an in-house developed 39-AIM-InDel panel and further admixture proportion estimation for Uyghurs. Electrophoresis. 2019. https://doi.org/10.1002/elps.201800448.
Li CT, Zhao SM, Zhang SH, Li L, Liu Y, Chen JZ, et al. Genetic polymorphism of 29 highly informative InDel markers for forensic use in the Chinese Han population. Forensic Sci Int-Gen. 2011;5:E27–E30.
Larue BL, Lagace R, Chang CW, Holt A, Hennessy L, Ge J, et al. Characterization of 114 insertion/deletion (INDEL) polymorphisms, and selection for a global INDEL panel for human identification. Leg Med-Tokyo. 2014;16:26–32.
Zhu B, Lan Q, Guo Y, Xie T, Fang Y, Jin X, et al. Population genetic diversity and clustering analysis for chinese dongxiang group with 30 autosomal indel loci simultaneously analyzed. Front Genet. 2018;9:279.
Wang L, Lv ML, Zaumsegel D, Zhang LS, Liu FJ, Xiang JH, et al. A comparative study of insertion/deletion polymorphisms applied among Southwest, South and Northwest Chinese populations using Investigator (R) DIPplex. Forensic Sci Int-Gen. 2016;21:10–14.
Guo Y, Shen C, Meng H, Dong Q, Kong T, Yang C, et al. Population differentiations and phylogenetic analysis of tibet and Qinghai Tibetan Groups based on 30 indel loci. DNA Cell Biol. 2016;35:787–94.
Kong T, Chen Y, Guo Y, Wei Y, Jin X, Xie T, et al. Autosomal InDel polymorphisms for population genetic structure and differentiation analysis of Chinese Kazak ethnic group. Oncotarget. 2017;8:56651–58.
Hong L, Wang XG, Liu SJ, Zhang YM, Xue-Ling OU, Chen Y, et al. Genetic polymorphisms of 30 indel loci in guangdong han population. J Sun Yat-Sen Univ (Med Sci). 2013;34:299–304.
Guo Y, Chen C, Jin X, Cui W, Wei Y, Wang H, et al. Autosomal DIPs for population genetic structure and differentiation analyses of Chinese Xinjiang Kyrgyz ethnic group. Sci Rep. 2018;8:11054.
Zhang YD, Shen CM, Jin R, Li YN, Wang B, Ma LX, et al. Forensic evaluation and population genetic study of 30 insertion/deletion polymorphisms in a Chinese Yi group. Electrophoresis. 2015;36:1196–201.
Wang Z, Zhang S, Zhao S, Hu Z, Sun K, Li C. Population genetics of 30 insertion-deletion polymorphisms in two Chinese populations using Qiagen Investigator(R) DIPplex kit. Forensic Sci Int Genet. 2014;11:e12–4.
Shi M, Liu Y, Bai R, Jiang L, Lv X, Ma S. Population data of 30 insertion-deletion markers in four Chinese populations. Int J Leg Med. 2015;129:53–6.
Shen C, Zhu B, Yao T, Li Z, Zhang Y, Yan J, et al. A 30-InDel assay for genetic variation and population structure analysis of Chinese Tujia Group. Sci Rep. 2016;6:36842.
Mei T, Shen CM, Liu YS, Meng HT, Zhang YD, Guo YX, et al. Population genetic structure analysis and forensic evaluation of Xinjiang Uigur ethnic group on genomic deletion and insertion polymorphisms. Springerplus.2016;5:1087.
Ma R, Shen C, Wei Y, Jin X, Guo Y, Mu Y, et al. Genetic differentiation and forensic efficiency evaluation for Chinese Salar ethnic minority based on a 5-dye multiplex insertion and deletion panel. Gene. 2018;660:41–50.
Meng HT, Zhang YD, Shen CM, Yuan GL, Yang CH, Jin R, et al. Genetic polymorphism analyses of 30 InDels in Chinese Xibe ethnic group and its population genetic differentiations with other groups. Sci Rep. 2015;5:8260.
Wei YL, Qin CJ, Dong H, Jia J, Li CX. A validation study of a multiplex INDEL assay for forensic use in four Chinese populations. Forensic Sci Int Genet. 2014;9:e22–5.
Chen YJ, Chen F, Xin N, Zhang HB, Zheng HB, Yu B, et al. [Genetic polymorphisms of X-STR loci in Chinese Yugur ethnic group and its application]. Yi Chuan. 2008;30:1143–52.
Yang F, An Y. Elements of Tibetan, Mongolian, the Tus and Formation of the Yugurs. Natl Res Qinghai. 2016;2:107–10.
Chen T, Jin TB, Xin N, Lai JH, Li SB. [Genetic polymorphisms and application of 9 STR loci of 5 ethnic groups in Gansu and Qinghai]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2006;31:877–82.
Yao Y, Shi L, Tao Y, Kulski JK, Lin K, Huang X, et al. Distinct HLA allele and haplotype distributions in four ethnic groups of China. Tissue Antigens. 2012;80:452–61.
Martin P, Garcia O, Heinrichs B, Yurrebaso I, Aguirre A, Alonso A. Population genetic data of 30 autosomal indels in Central Spain and the Basque Country populations. Forensic Sci Int-Gen. 2013;7:E27–E30.
Friis SL, Borsting C, Rockenbauer E, Poulsen L, Fredslund SF, Tomas C, et al. Typing of 30 insertion/deletions in Danes using the first commercial indel kit–Mentype(R) DIPplex. Forensic Sci Int Genet. 2012;6:e72–4.
Kis Z, Zalan A, Volgyi A, Kozma Z, Domjan L, Pamjav H. Genome deletion and insertion polymorphisms (DIPs) in the Hungarian population. Forensic Sci Int Genet. 2012;6:e125–6.
Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques. 1991;10:506–13.
Hill CR, Duewer DL, Kline MC, Sprecher CJ, McLaren RS, Rabbach DR, et al. Concordance and population studies along with stutter and peak height ratio analysis for the PowerPlex (R) ESX 17 and ESI 17 Systems. Forensic Sci Int Genet. 2011;5:269–75.
Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–7.
Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–8.
Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Computing. 2009;14:12–21.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59.
Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–8.
Yoo J, Lee Y, Kim Y, Rha SY, Kim Y. SNPAnalyzer 2.0: a web-based integrated workbench for linkage disequilibrium analysis and association analysis. BMC Bioinform. 2008;9:290.
Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic-linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–31.
Liu C, Liu C, Wang H. STR data for the 15 loci from three minority populations in Guangxi municipality in South China. Forensic Sci Int. 2006;162:49–52.
Zhang L, Zhao Y, Guo F, Liu Y, Wang B. Population data for 15 autosomal STR loci in the Miao ethnic minority from Guizhou Province, Southwest China. Forensic Sci Int Genet. 2015;16:e3–e4.
An Y. A Study in The Formation History of Yugurs. Lanzhou University; 2016.
Deng Q, Xu L, Gong J, Zhou L, Li S, Deng X, et al. Genetic relationships among four minorities in Guangxi revealed by analysis of 15 STRs. J Genet Genom. 2007;34:1072–9.
Zhang X, Hu L, Du L, Nie A, Rao M, Pang JB, et al. Genetic analysis of 20 autosomal STR loci in the Miao ethnic group from Yunnan Province, Southwest China. Forensic Sci Int Genet. 2017;28:e28–e29.
Acknowledgements
This study was supported by Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (GDUPS, 2017).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yang, CH., Jin, XY., Guo, YX. et al. Genetic distribution analyses and population background explorations of Gansu Yugur and Guizhou Miao groups via InDel markers. J Hum Genet 64, 535–543 (2019). https://doi.org/10.1038/s10038-019-0595-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0595-3