Abstract
Distal spinal muscular atrophy (dSMA) is a rare clinically and genetically heterogeneous group of inherited disorders characterized by progressive distal muscle weakness and wasting. So far, more than 65% of patients with dSMA have undiscovered genetic mutations. Recently, compound heterozygous mutations in the vaccinia-related kinase 1 (VRK1) gene have been identified for the first time in two siblings with adult-onset dSMA from an Ashkenazi Jewish family. Here, we also report two affected siblings with adult-onset dSMA in a Chinese family. Whole exome sequencing and subsequent Sanger sequencing identified a novel nonsense mutation (c.1124G >A, p.W375*) in exon 12 of the VRK1 gene, co-segregating with the dSMA phenotype in an autosomal recessive pattern. In conclusion, our findings identify a novel nonsense mutation p.W375* in the VRK1 gene in a Chinese family with autosomal recessive dSMA and broaden the genetic spectrum of VRK1-associated dSMA.
Similar content being viewed by others
Introduction
Distal spinal muscular atrophy (dSMA), also called distal hereditary motor neuropathy (dHMN), is a rare genetically and clinically heterogeneous group of disorders caused by degeneration and loss of anterior horn cells in the spinal cord, is characterized by slowly progressive distal muscle weakness and atrophy without sensory abnormalities [1]. Nevertheless, many forms of dSMA have other additional manifestations including minor sensory abnormalities and pyramidal signs, with overlap among spinal muscular atrophy (SMA), and juvenile amyotrophic lateral sclerosis (ALS), hereditary motor and sensory neuropathy (HMSN), hereditary spastic paraplegia (HSP), where mutations in the same gene may even cause different phenotypes [2]. To date, at least 20 genes and 4 loci associated with dSMA have been reported (http://neuromuscular.wustl.edu/). However, more than 65% of patients with dSMA have undiscovered genetic mutations [3].
Initially, a nonsense mutation c.1072C>T (p.R358*) in the vaccinia-related kinase 1 (VRK1) gene was found as a cause of infantile-onset spinal muscular atrophy with pontocerebellar hypoplasia (SMA-PCH) in a consanguineous Ashkenazi Jewish family [4]. Subsequently, the same homozygous mutation p.R358* and pathogenic compound heterozygous mutations (c.706G>A, p.V236M and c.266G>A, p.R89Q) in VRK1 were also associated with HMSN accompanied by microcephaly and cerebral dysgenesis [5]. Moreover, additional compound heterozygous mutations in the VRK1 gene have also been identified in early-onset and juvenile ALS [6, 7].
Recently, compound heterozygous VRK1 mutations, p.R358* and c.356A>G (p.H119R), have been identified for the first time in two siblings with adult-onset dSMA from a family of Ashkenazi Jewish origin [7]. In the present study, we also report two affected siblings with adult-onset dSMA caused by a novel nonsense mutation in the VRK1 gene in a Chinese family.
Materials and Methods
Subjects
A Chinese family with two affected siblings participated in our study. Nine family members including the affected individuals were comprehensively checked by two experienced neurologists. All individuals in the study signed informed written consent before enrollment. This study was approved by the Ethics Committee of Sichuan University.
Genetic analysis
DNA was extracted from blood leukocytes by standard procedures. Initially, the deletion of the survival motor neuron 1 (SMN1) gene associated with SMA was excluded in the proband. To identify the causative variant in this family, we completed whole exome sequencing of the proband (II:7), his affected sister (II:9) and their sister (II: 5). For exome capture, Agilent SureSelect Human All Exon v6 Kit (Agilent, Santa Clara, CA, USA) was used. Captured libraries were constructed as instructed by the manufacturer’s protocols. Sequencing was carried out by the Illumina HiSeq X10 platform (Illumina, San Diego, CA, USA) as paired-end 150-bp reads. Quality control was applied to raw data to guarantee the meaningfulness of downstream analysis. Sequence reads were analyzed and aligned to the human reference genome (hg19) using Burrows-Wheeler Alignment tool. The variants were discovered and annotated by Genome Analysis Toolkit (GATK) and ANNOVAR. All variants were first filtered against 1000 genomes project database (http://www.internationalgenome.org/), and variants with a minor allele frequency (MAF) ≥ 1% were removed from the analysis. Variants were further filtered according to co-segregation with the disease and a MAF < 1% in public variant databases. Only variants occurring in exons and canonical splice sites were further analyzed. Subsequently, Sanger sequencing was performed to verify the putative mutation in all available family members. The following primers were designed for polymerase chain reaction (PCR) amplification: 5ʹ-GTGTTTTGACACACCCACACC-3ʹ (forward) and 5ʹ-CACATCAGTCACCACGCCT-3ʹ (reverse). Additionally, the pathogenicity of the potential causative variation was predicted using MutationTaster (http://www.mutationtaster.org).
Results
Clinical features
Two affected siblings were from a nonconsanguineous family of Chinese Han origin (Fig. 1a). The 46-year-old proband (II:7) had difficulties in walking on tiptoe from the age of 13 years. He developed a slowly progressive muscle atrophy of both feet and calves at age 17 years. Subsequently, progressive distal wasting and weakness of the upper limbs became noted at age 26 years. He had difficulties in climbing the stairs without support at age 30 years. The impairment of motor function was progressive over time and at age 42 years he required a wheelchair to mobilize. He denied dysphagia, dysarthria, dyspnea, sensory symptoms, or sphincter disturbance across the course of the illness and he had normal intellectual development.
Pedigree of the Chinese family with dSMA and the c.1124G>A mutation of VRK1. a Pedigree of the Chinese family with dSMA. Black boxes indicate the patients affected. The arrow indicates the proband (patient II:7). b Chromatograms of the VRK1 gene. The patient and the carrier harbored the homozygous and heterozygous c.1124G>A mutation, respectively (arrows)
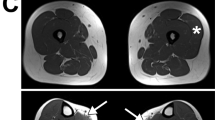
Neurological examination revealed normal mental status and cranial nerves. He had symmetrical and predominantly distal muscle wasting and weakness of greater severity in the lower limbs (Medical Research Council [MRC] grade 0/5 distal and 2/5 proximal) than in the upper limbs (MRC grade 3/5 distal and 4/5 proximal) (Fig. 2), hypotonia in all limbs, and absence of fasciculation and deep tendon reflexes. There were no sensory abnormalities, pyramidal signs, or cerebellar ataxia. Serum creatine kinase (CK) was at normal level. Neurophysiology showed severe reduction in compound muscle action potential amplitudes with preservation of motor nerve conduction velocities, and normal sensory nerve conductions (Supplementary Table 1).
Electromyography (EMG) showed chronic neurogenic changes of high amplitude, long duration motor units. Magnetic resonance imaging (MRI) of both brain and spinal cord at age 36 years was normal and did not show pontine or cerebellar hypoplasia (Images are unavailable). In addition, he refused to accomplish the nerve and muscle biopsy.
The proband has an affected sister (II:9) with similar clinical symptoms. She presented with progressive wasting and weakness of distal symmetrical lower limbs from the age of 16 years. She had difficulties climbing the stairs without support at age 30 years. She walked with a high steppage gait due to bilateral foot drop at age 40 years. At age 41 years, she was found right cerebellar hemangioblastoma treated with complete surgical resection. Her neurological examination showed symmetrically severe and moderate wasting and weakness in lower extremities (MRC grade 1/5 distal and 4/5 proximal), slight wasting and weakness of the ulnar intrinsic muscle of both hands, pes arcuatus, hypotonia in all limbs. Notably, deep tendon reflexes were also absent in the lower limbs but symmetrically brisk in the upper limbs. The rest of the neurologic examination was normal. Serum CK was mildly elevated (166 IU/L, normal range 20–140 IU/L). A reduction in compound muscle action potential of the peroneal nerve was observed (Supplementary Table 1). The EMG also revealed chronic neurogenic changes. The right quadriceps muscle biopsy showed marked small angular fibers and pyknotic nuclear clumps and large-group atrophy with increased connective tissue that was consistent with neurogenic atrophy. Furthermore, the brain and whole spinal MRI were normal except for surgical resection of the right cerebellar hemangioblastoma.
The two affected siblings showed no delay in developmental milestone achievements. Surprisingly, the other two members of the family also suffered from tumors except the affected sister. Their oldest sister (II:1) was diagnosed with kidney cancer at age 37 years. Subsequently, she (II:9) was also found cerebellar hemangioblastoma. Their older sister (II:3) was diagnosed with advanced liver cancer and died at age 53 years.
Identification of homozygous nonsense mutation in VRK1
In the VRK1 gene, the proband was found to have a novel homozygous nonsense mutation c.1124G>A of VRK1 exon 12 (NM_003384.2), producing a premature stop codon (p.W375*). The affected sister also harbored the same homozygous nonsense mutation p.W375*. The novel nonsense mutation in the VRK1 gene completely co-segregated with the disease phenotype in an autosomal recessive pattern in all available family members (Fig. 1). The p.W375* is absent in the Exome Aggregation Consortium databases and is a very low MAF (A = 0.0002/1) in 1000 Genome project databases. Moreover, the novel nonsense mutation p.W375* was predicted to be disease-causing by MutationTaster, with a probability value of >0.9999.
Discussion
The VRK1 gene is widely expressed and elevated in highly proliferative cells such as fetal liver, testis, and thymus [8]. This gene comprises 13 exons and encodes a 396 aa protein (NP_003375), with an N-terminal serine-threonine kinase domain, a C-terminal nuclear location signal (NLS), an endosomal and lysosomal targeting sequence, and a basic-acidic-basic (BAB) motif at the C terminus. The VRK1 protein is a nuclear serine-threonine protein kinase and has been most extensively studied in cellular proliferation, tumorigenesis, and DNA damage response [9]. Recently, several studies have showed that VRK1 is involved in several tumors, such as hepatocellular carcinoma [10], lung carcinomas [11], glioblastoma [12], breast cancer [13], and so on. Surprisingly, one homozygous (II:9) and two heterozygous p.W375* carriers (II:1 and II:3) of the family also suffered from tumors. But, functional studies of the nonsense mutation p.W375* in the VRK1 gene to unravel its role in tumorigenesis will be useful.
Interestingly, the nonsense mutation p.R358* in the VRK1 gene seems to be a rare founder mutation in the Ashkenazi Jewish population4. The p.R358* within NLS of VRK1 disturbs its subcellular localization and the truncated VRK1 (p.R358*) protein does not properly localize to the nucleus but localize to the cytoplasm [14]. Moreover, the C-terminal region of VRK1 has a flexible conformation that modulates kinase activity and the progressive deletion of the C-terminal tail resulted in a gradual reduction in the autocatalytic activity of VRK1 [15]. In vitro kinase assay, the kinase activity of truncated VRK1 (p.R358*) protein, lacking the terminal 38 amino acids, is proximately eightfold less than wild-type VRK1 [14]. However, since the p.R358* transcript undergoes nonsense-mediated mRNA decay (NMD), the truncated VRK1 (p.R358*) was not detectable in homozygous patients with SMA-PCH [4, 16].
In this study, the results are consistent with an autosomal recessive pattern with co-segregation of the novel nonsense mutation p.W375* in the VRK1 gene with adult-onset dSMA. The p.W375* variant encodes a tryptophan residue that is relatively conserved across multiple vertebrate species and is in the BAB motif of the unknown function near the NLS. The truncated VRK1 (p.W375*) protein lacks the terminal 21 amino acids of C-terminal region of VRK1. This suggests that the truncated VRK1 (p.W375*) protein might also alter its subcellular localization and decrease its kinase activity like truncated VRK1 (p.R358*). Importantly, the homozygous nonsense mutation p.W375* in the VRK1 gene has been found to cause the clinical phenotypes of dSMA in this study [17], which is nearly completely consistent with the phenotypes of p.H119R and p.R358* compound heterozygous mutations in Ashkenazi Jewish family [7]. However, it is still unknown whether the novel p.W375* transcript also is degraded by triggering NMD pathway.
The latest study shows that VRK1 knockdown significantly impairs embryonic cortical neuronal migration and affects the cell cycle of neuronal progenitors, which provides the first insight into the molecular mechanism of VRK1-related neurologic disease [16]. However, it is difficult to explain why VRK1 mutations result in pathology restricted to spinal motor neurons in our patients. Notably, dysfunction of Cajal bodies (CBs) associated with ribonucleoprotein functions and RNA maturation could be involved in SMA pathophysiology [18, 19]. Recently, VRK1 has been shown to interact with and phosphorylate coilin in Ser184, a major structural scaffolding protein of CBs, regulating assembly and stability of CBs in cell cycle [20]. Therefore, these might provide clues to the role of VRK1 in the pathogenesis of dSMA. More functional studies are required to unravel the role of VRK1 in the nervous system and address the biological consequences of mutations in patients.
In summary, we identified a novel nonsense mutation p.W375* in the VRK1 gene in an extremely rare Chinese family with autosomal recessive dSMA. Our findings broaden the genetic spectrum of VRK1-associated dSMA and have implications for molecular diagnostics of dSMA.
References
Harding AE. Inherited neuronal atrophy and degeneration predominantly of lower motor neurons. In: Dyck PJ, Thomas PK (eds). Peripheral Neuropathy, 4th edn. pp. 1603–21. WB Saunders: Philadelphia, PA, USA, 2005.
Rossor AM, Kalmar B, Greensmith L, Reilly MM. The distal hereditary motor neuropathies. J Neurol Neurosurg Psychiatry. 2012;83:6–14.
Bansagi B, Griffin H, Whittaker RG, Antoniadi T, Evangelista T, Miller J, et al. Genetic heterogeneity of motor neuropathies. Neurology. 2017;88:1226–34.
Renbaum P, Kellerman E, Jaron R, Geiger D, Segel R, Lee M, et al. Spinal muscular atrophy with pontocerebellar hypoplasia is caused by a mutation in the VRK1 gene. Am J Hum Genet. 2009;85:281–9.
Gonzaga-Jauregui C, Lotze T, Jamal L, Penney S, Campbell IM, Pehlivan D, et al. Mutations in VRK1 associated with complex motor and sensory axonal neuropathy plus microcephaly. JAMA Neurol. 2013;70:1491–8.
Nguyen TP, Biliciler S, Wiszniewski W, Sheikh K. Expanding phenotype of VRK1 mutations in motor neuron disease. J Clin Neuromuscul Dis. 2015;17:69–71.
Stoll M, Teoh H, Lee J, Reddel S, Zhu Y, Buckley M, et al. Novel motor phenotypes in patients with VRK1 mutations without pontocerebellar hypoplasia. Neurology. 2016;87:65–70.
Nezu J, Oku A, Jones MH, Shimane M. Identification of two novel human putative serine/threonine kinases, VRK1 and VRK2, with structural similarity to vaccinia virus B1R kinase. Genomics. 1997;45:327–31.
Valbuena A, Sanz-Garcia M, Lopez-Sanchez I, Vega FM, Lazo PA. Roles of VRK1 as a new player in the control of biological processes required for cell division. Cell Signal. 2011;23:1267–72.
Lee N, Kwon JH, Kim YB, Kim SH, Park SJ, Xu W, et al. Vaccinia-related kinase 1 promotes hepatocellular carcinoma by controlling the levels of cell cycle regulators associated with G1/S transition. Oncotarget. 2015;6:30130–48.
Valbuena A, Suarez-Gauthier A, Lopez-Rios F, Lopez-Encuentra A, Blanco S, Fernandez PL, et al. Alteration of the VRK1-p53 autoregulatory loop in human lung carcinomas. Lung Cancer. 2007;58:303–9.
Varghese RT, Liang Y, Guan T, Franck CT, Kelly DF, Sheng Z. Survival kinase genes present prognostic significance in glioblastoma. Oncotarget. 2016;7:20140–51.
Salzano M, Vazquez-Cedeira M, Sanz-Garcia M, Valbuena A, Blanco S, Fernandez IF, et al. Vaccinia-related kinase 1 (VRK1) confers resistance to DNA-damaging agents in human breast cancer by affecting DNA damage response. Oncotarget. 2014;5:1770–8.
Sanz-Garcia M, Vazquez-Cedeira M, Kellerman E, Renbaum P, Levy-Lahad E, Lazo PA. Substrate profiling of human vaccinia-related kinases identifies coilin, a Cajal body nuclear protein, as a phosphorylation target with neurological implications. J Proteom. 2011;75:548–60.
Shin J, Chakraborty G, Bharatham N, Kang C, Tochio N, Koshiba S, et al. NMR solution structure of human vaccinia-related kinase 1 (VRK1) reveals the C-terminal tail essential for its structural stability and autocatalytic activity. J Biol Chem. 2011;286:22131–8.
Vinograd-Byk H, Sapir T, Cantarero L, Lazo PA, Zeligson S, Lev D, et al. The spinal muscular atrophy with pontocerebellar hypoplasia gene VRK1 regulates neuronal migration through an amyloid-beta precursor protein-dependent mechanism. J Neurosci. 2015;35:936–42.
2nd Workshop of the European CMT Consortium: 53rd ENMC International Workshop on Classification and Diagnostic Guidelines for Charcot-Marie-Tooth Type 2 (CMT2-HMSN II) and Distal Hereditary Motor Neuropathy (distal HMN-Spinal CMT) 26-28 September 1997, Naarden, The Netherlands. Neuromuscul Disord 1998;8:426–31.
Tapia O, Bengoechea R, Palanca A, Arteaga R, Val-Bernal JF, Tizzano EF, et al. Reorganization of Cajal bodies and nucleolar targeting of coilin in motor neurons of type I spinal muscular atrophy. Histochem Cell Biol. 2012;137:657–67.
Tapia O, Lafarga V, Bengoechea R, Palanca A, Lafarga M, Berciano MT. The SMN Tudor SIM-like domain is key to SmD1 and coilin interactions and to Cajal body biogenesis. J Cell Sci. 2014;127(Pt 5):939–46.
Cantarero L, Sanz-Garcia M, Vinograd-Byk H, Renbaum P, Levy-Lahad E, Lazo PA. VRK1 regulates Cajal body dynamics and protects coilin from proteasomal degradation in cell cycle. Sci Rep. 2015;5:10543.
Acknowledgements
The authors are thankful to the patients and their families for their participation in the present study.
Funding
This study was supported by the China Postdoctoral Science Foundation (Number: 2017M610605) and Sichuan Science and Technology Program (Number: 2018FZ0029).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, N., Wang, L., Sun, X. et al. A novel mutation in VRK1 associated with distal spinal muscular atrophy. J Hum Genet 64, 215–219 (2019). https://doi.org/10.1038/s10038-018-0553-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-018-0553-5