Abstract
Our original microRNA (miRNA) expression signatures (based on RNA sequencing) revealed that both strands of the miR-145 duplex (miR-145-5p, the guide strand, and miR-145-3p, the passenger strand) were downregulated in several types of cancer tissues. Involvement of passenger strands of miRNAs in cancer pathogenesis is a new concept in miRNA biogenesis. In our continuing analysis of lung adenocarcinoma (LUAD) pathogenesis, we aimed here to identify important oncogenes that were controlled by miR-145-5p and miR-145-3p. Downregulation of miR-145-5p and miR-145-3p was confirmed in LUAD clinical specimens. Functional assays showed that miR-145-3p significantly blocked the malignant abilities in LUAD cells, e.g., cancer cell proliferation, migration and invasion. Thus, the data showed that expression of the passenger strand of the miR-145-duplex acted as an anti-tumor miRNA. In LUAD cells, we identified four possible target genes (LMNB2, NLN, SIX4, and DDC) that might be regulated by both strands of miR-145. Among the possible targets, high expression of LMNB2 predicted a significantly poorer prognosis of LUAD patients (disease-free survival, p = 0.0353 and overall survival, p = 0.0017). Overexpression of LMNB2 was detected in LUAD clinical specimens and its aberrant expression promoted malignant transformation of LUAD cells. Genes regulated by anti-tumor miR-145-5p and miR-145-3p are closely involved in the molecular pathogenesis of LUAD. We suggest that they are promising prognostic markers for this disease. Our approach, based on the roles of anti-tumor miRNAs, will contribute to improved understanding of the molecular pathogenesis of LUAD.
Similar content being viewed by others
Introduction
Lung cancer is the leading cause of cancer-related deaths around the world. In 2015, approximately 2.2 million individuals were diagnosed with lung cancer, resulting in the death of 1.7 million patients [1]. Non-small cell lung cancer (NSCLC) is the most common type of lung cancer (approximately 80–85% of this disease). It is divided into three subtypes: squamous cell carcinoma (LUSQ), adenocarcinoma (LUAD), and large cell carcinoma [2]. About 40% of lung cancers are LUAD, and this type tends to grow more slowly than other types of lung cancer [3]. There are currently numerous treatment options available for patients with LUAD depending on the stage of the cancer, the patient’s overall health and their cancer genomic status [4]. Due to recently developed molecularly targeted drugs and immune checkpoint inhibitors, the treatment outcome of patients with LUAD has been markedly improved. However, the therapeutic effectiveness for patients with distant metastasis is limited and the prognosis is extremely poor. To develop effective therapies, genomic studies of LUAD are being conducted worldwide using the latest genome analysis technology.
microRNAs (miRNAs) classified as small non-coding RNAs exist in the human genome and have been actually transcribed. They regulate the expression of protein coding/non-coding RNAs by repressing their translation or by cleaving transcripts. This is achieved in a sequence-dependent manner [5]. A single miRNA species can regulate a large number of RNA transcripts in physiological and pathological conditions [5]. Therefore, aberrantly expressed miRNAs can disrupt RNA and protein populations critical for normal cell maintenance and growth [6]. In fact, many studies have shown that dysregulated expression of miRNAs enhances cancer cell development, metastasis, and drug resistance [7]. Thus, aberrantly expressed miRNA can be closely involved in human cancer pathogenesis [8].
We have used RNA sequencing techniques to obtain miRNA expression signatures in cancer tissues. The data revealed that the expression of some miRNA passenger strands was upregulated or downregulated, suggesting that certain passenger strands might be involved in cancer pathogenesis [9, 10]. Our recent studies revealed that some passenger strands (miR-145-3p, miR-150-3p, miR-199a-3p, miR-216b-3p, and miR-99a-3p) targeted oncogenes in several cancers [11,12,13,14,15,16]. In fact, our results showed that both strands of the miR-145 duplex (miR-145-5p, the guide strand and miR-145-3p, the passenger strand) possess anti-tumor functions in LUSQ cells and bladder cancer [17, 18]. However, the functional significance of miR-145-3p and its targets in LUAD cells is still obscure.
In our continuing analysis of LUAD pathogenesis, we aimed here to identify important oncogenes that are controlled by miR-145-5p and miR-145-3p. We identified four possible target genes (LMNB2, NLN, SIX4, and DDC) that might be regulated by both strands of the miR-145 duplex. Among the targets, high expression of LMNB2 was significantly associated with poor prognosis of the patients with LUAD (disease-free survival, p = 0.0353 and overall survival, p = 0.0017). Overexpression of LMNB2 was observed in LUAD clinical specimens and its expression enhanced cancer cell aggressiveness. Defining the targets of anti-tumor miRNAs will enhance our understanding of the molecular pathology of LUAD. Moreover, these insights could lead to the development of early detection markers of LUAD and new therapeutic drugs.
Materials and methods
Human LUAD specimens
Formalin-fixed, paraffin-embedded specimens (FFPE) (19 LUAD tissues and 24 normal lung tissues) were collected at Kagoshima University Hospital (from 2010 to 2013) where patients underwent lung surgery. The clinical features of these specimens are summarized in Supplemental Table 1. The pathological stages of these LUAD patients were determined according to the Association for the Study of Lung Cancer TNM classification, 7th edition.
Tissue samples for this study were obtained with informed consent and approval from each patient. Our study was approved by the Institutional Review Board for Clinical Research of Kagoshima University Hospital (approval number 26-164).
RNA extraction, reverse transcription, and quantitative real-time PCR
RNA extraction from FFPE specimens and cell lines and quantitative real-time reverse transcription-PCR (qRT-PCR) were described in our previous studies [10, 19, 20].
Expression levels of miR-145-5p and miR-145-3p were analyzed by stem-loop RT-PCR (assay ID: 002278 and 002149; Applied Biosystems). They were normalized to RNU48 (assay ID: 001006; Applied Biosystems). TaqMan probes and primers for LMNB2 (P/N: Hs00383326_m1) and GUSB (P/N: Hs00939627_m1) as an internal control were obtained from Applied Biosystems.
Cell culture and transfection of miRNAs and siRNAs
Two human LUAD cell lines (A549 and H1299) were used in this study. Both were purchased from the ATCC (Manassas, VA, USA). The procedures for transfection of miRNA or siRNA species into cancer cells were described in our recent reports [20,21,22].
The following miRNA species were used in this study: Pre-miRTM miRNA precursors (hsa-miR-145-5p, assay ID: PM 11480; hsa-miR-145-3p, assay ID: PM 13036; negative control miRNA, catalog no: 4464059, AM17111; Applied Biosystems). Also, we obtained the following siRNAs from Invitrogen (Carlsbad, CA, USA): Stealth Select RNAi siRNA, si-LMNB2 (P/N: HSS189213 and HSS189214; Invitrogen), and negative control miRNA/siRNA, Anti-miR Negative Control #1 (catalog no: AM 17010; Applied Biosystems).
Incorporation of miR-145-5p or miR-145-3p into the RISC by Ago2 immunoprecipitation
To determine whether exogenous miR-145-5p and miR-145-3p were incorporated into the RISC, immunoprecipitation assays were performed as described previously [14, 15, 23].
Cell proliferation, migration and invasion assays
The procedures for assessing cell proliferation, migration and invasion were described previously [19,20,21].
Identification of putative target genes regulated by miR-145-5p and miR-145-3p in LUAD cells
Microarray gene expression analysis and in silico analysis were used to identify putative target genes regulated by miR-145-5p and miR-145-3p in LUAD cells. The microarray data were deposited in the Gene Expression Omnibus (GEO) repository under accession number GSE77790. Our strategy for identification of miR-145-5p and miR-145-3p target genes is outlined in Supplemental Fig. 1.
Plasmid construction and dual-luciferase reporter assay
The wild-type or deletion-type sequences targeted by miR-145-5p or miR-145-3p were inserted in the psiCHECK-2 vector (C8021; Promega, Madison, WI, USA).
After co-transfecting miRNA and the constructed vector in A549 and H1299 cells, firefly and Renilla luciferase activities were measured using a dual-luciferase assay kit (Promega Madison, WI, USA). The procedure was described previously [17, 20, 24].
Clinical database analysis
The clinical significance of miRNAs and their target genes in LUAD was investigated with TCGA database (The Cancer Genome Atlas: https://tcga-data.nci.nih.gov/tcga/). The gene expression and clinical data were retrieved from cBioPortal (http://www.cbioportal.org/) and OncoLnc (data downloaded on April 24, 2018) [25]. The normalized mRNA expression values in the RNA sequencing data were processed and shown as Z-scores. In the present study, patients were divided into two groups: those with Z-score ≥ 0 and those with Z-score < 0. The TNM stage of LUAD patients was determined by the American Joint Committee on Cancer staging system in TCGA database.
Western blotting and immunohistochemistry
The procedures for Western blotting and immunohistochemistry were described previously [19, 20, 24]. Membranes were immunoblotted with monoclonal anti-LMNB2 antibody (1:1000 dilution) (ab8983; Abcam, Cambridge, UK) and monoclonal anti-GAPDH antibody (1:10,000 dilution) (MAB374; EMD Millipore, Billerica, MA, USA).
Immunohistochemistry was performed with a VECTASTAIN® Universal Elite ABC Kit (catalog no: PK-6200; Vector Laboratories, Burlingame, CA, USA) according to the manufacturer’s protocol. Primary mouse monoclonal antibody against LMNB2 (ab8983; Abcam) was used at a 1:100 dilution at 4 °C overnight.
Statistical analysis
Mann–Whitney U tests were used to analyze the differences between two groups. Bonferroni-adjusted Mann–Whitney U tests were used to analyze the differences among more than three groups. Statistical tests were performed using Expert StatView software (version 5.0, SAS Institute Inc., Cary, NC, USA). Survival analysis was performed using the Kaplan–Meier method, and log-rank tests and multivariable Cox hazard regression analyses with JMP software (version 14; SAS Institute Inc., Cary, NC, USA).
Results
Downregulation of miR-145-5p and miR-145-3p in LUAD clinical specimens and cell lines
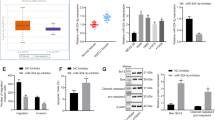
The expression levels of miR-145-5p and miR-145-3p were remarkably decreased in LUAD tissues in comparison with non-cancerous tissues (p < 0.0001 and p = 0.0039, respectively, Fig. 1a, b). Positive correlations between the expression of miR-145-5p and miR-145-3p were confirmed by Spearman’s rank test (R = 0.862 and p < 0.0001, Fig. 1c). In cancer cell lines, A549 and H1299, the expression levels of miR-145-5p and miR-145-3p were extremely low (Fig. 1a, b). There was no correlation between expression levels of miR-145-5p and miR-145-3p, and clinical features of LUAD (Supplemental Fig. 2).
The expression levels of miR-145-5p and miR-145-3p in lung adenocarcinoma (LUAD) clinical specimens and functional assays of miR-145-5p-transfected and miR-145-3p-transfected LUAD cell lines. a, b Expression levels of miR-145-5p and miR-145-3p in clinical specimens (LUAD and non-cancerous tissue) and LUAD cell lines (A549 and H1299) were determined by quantitative real-time reverse transcription-PCR (qRT-PCR). Data were normalized to RNU48 expression. The expression levels of miR-145-5p and miR-145-3p in cancer tissues were significantly downregulated in comparison with non-cancerous tissues (p < 0.0001, p = 0.0039, respectively). c Spearman’s rank test indicated a positive correlation between the expression of miR-145-5p and miR-145-3p (R = 0.862 and p < 0.0001). d Cell proliferation was evaluated by XTT assays. e Cell migration was determined by wound healing assay. f Cell invasion was determined by Matrigel invasion assay. Inhibition of proliferation, migration, and invasion was found in miR-145-5p-transfected and miR-145-3p-transfected cell lines (p < 0.0001). *p < 0.0001, **p < 0.05
Ectopic expression of miR-145-5p and miR-145-3p suppressed cancer cell proliferation, migration, and invasive abilities
To verify the anti-tumor roles of miR-145-5p and miR-145-3p, we conducted gain-of-function studies by miRNA transfection into A549 and H1299 cells. Cell proliferation assays indicated the inhibition of cancer cell growth in miR-145-5p and miR-145-3p-transfected cells compared with mock-transfected or miR-control-transfected cells (both p < 0.0001, Fig. 1d). Also, cell migration activities were reduced in the cells transfected with miR-145-5p or miR-145-3p (both p < 0.0001, Fig. 1e). Moreover, Matrigel invasion assays revealed that transfection with miR-145-5p and miR-145-3p remarkedly decreased cell invasive capacity (both p < 0.0001, Fig. 1f).
Identification of putative target genes regulated by miR-145-5p and miR-145-3p in LUAD cells: clinical significance of LMNB2, NLN, SIX4, and DDC
Using TargetScan database analysis, there were a total of 3773 and 2727 putative target genes that had binding sites for miR-145-5p and miR-145-3p, respectively. Among them, we selected genes that were upregulated in NSCLC clinical expression profiles from the GEO database (accession no: GSE19188). Then, we merged gene expression analysis data using miR-145-5p-transfected or miR-145-3p-transfected A549 cells (GEO accession number: GSE77790) and chose the downregulated genes. Finally, a total of four putative target genes (LMNB2, NLN, SIX4, and DDC) regulated by both miR-145-5p and miR-145-3p were identified (Tables 1A, 1B, 1C).
To investigate the relationship between these four genes and the course of LUAD, we examined these genes with the TCGA database (Fig. 2a–d). Among these candidates, high expression of LMNB2 was associated with poor prognosis (disease-free survival, p = 0.0353, and overall survival, p = 0.0017). In this study, we focused on LMNB2 and validated the effect on LUAD cells.
Clinical significance of LMNB2, NLN, SIX4, and DDC expression based on The Cancer Genome Atlas (TCGA) database. a–d The relationship between the expression of four genes (LMNB2, NLN, SIX4, and DDC) and disease-free survival analyses, overall survival analyses, T stage, N stage, M stage, and disease stage. **p < 0.05
Incorporation of miR-145-5p and miR-145-3p into the RISC in LUAD cells
We hypothesized that the miR-145-3p passenger strand might be incorporated into the RISC in LUAD cells. If so, it could serve as a tumor suppressor. To verify that concept, we immunoprecipitated Ago2 in cells that had been transfected with either miR-145-5p or with miR-145-3p. Note that Ago2 is an essential component of the RISC. Isolated Ago2-bound miRNAs were analyzed by qRT-PCR to determine whether miR-145-5p or miR-145-3p or both bound to Ago2 (Supplemental Fig. 3A). In A549 cells, miR-145-3p transfectants showed higher expression levels of miR-145-3p than did mock transfectants or miR-control (p < 0.0001). Similarly, after miR-145-5p transfection, miR-145-5p was detected by Ago2 immunoprecipitation (p < 0.0001) (Supplemental Fig. 3B).
Direct regulation of LMNB2 by miR-145-5p or miR-145-3p in LUAD cells
We examined whether the expression of LMNB2 mRNA and LMNB2 protein was suppressed by the restoration of miR-145-5p and miR-145-3p in LUAD cells. The expression levels of LMNB2 mRNA and LMNB2 protein were significantly decreased by miR-145-5p or miR-145-3p transfection compared with mock- or miR-control-transfected cells (Fig. 3a).
Both strands of miR-145 (miR-145-5p and miR-145-3p) directly regulated LMNB2 in LUAD cell lines. a Expression levels of LMNB2 mRNA and LMNB2 protein 48 h after transfection of 10 nM miR-145-5p or miR-145-3p into LUAD cell lines. GUSB and GAPDH was used as an internal control. *p < 0.0001. b, c miR-145-5p or miR-145-3p binding sites in the 3′-UTR of LMNB2. Luciferase assay using the vectors encoding putative miR-145-5p or miR-145-3p target sites of the LMNB2 3′-UTR for both wild-type and deletion-type constructs. Renilla luciferase values were normalized to firefly luciferase values. *p < 0.0001
Next, we sought to confirm that miR-145-5p and miR-145-3p directly regulated the LMNB2 gene in a sequence-dependent manner. TargetScan database projected the existence of binding sites in the 3′-UTR of LMNB2 (miR-145-5p, position 2700–2706; miR-145-3p, position 1689–1696, Fig. 3b, c). Accordingly, we carried out luciferase reporter assays with vectors that included either the wild-type or deletion-type 3′-UTR of LMNB2. The luminescence intensities were markedly reduced by transfection with miR-145-5p or miR-145-3p and the vector carrying the wild-type 3′-UTR of LMNB2. In contrast, transfection with the deletion-type vector did not reduce the luminescence intensities in LUAD cells (A549 and H1299) (Fig. 3b, c). These findings showed that both strands of the miR-145 duplex directly bound the 3′-UTR of LMNB2.
In human genome, miR-143 and miR-145 are known to be located close together on human chromosome 5q32 region, it called as the clustered miRNA. The TargetScan database search revealed that a putative binding site of miR-143-3p (the guide strand) has annotated in the 3′-UTR of LMNB2. We also investigated the regulation of LMNB2 by miR-143-3p in LUAD cells. The mRNA and protein expression levels of LMNB2 were markedly downregulated in miR-143-3p expression (Supplemental Fig. 4A, B). The luciferase reporter assay showed that miR-143-3p was directly bound to the 3′-UTR of LMNB2 (Supplemental Fig. 4C). These findings indicated that LMNB2 was directly regulated by miR-143-3p in LUAD cells.
LMNB2 knockdown inhibited cell proliferation, migration, and invasion in LUAD cells
To analyze the oncogenic role of LMNB2 in LUAD cells, loss-of-function assays were performed using si-LMNB2. We confirmed that both siRNAs (si-LMNB2-1 and si-LMNB2-2) significantly suppressed the expression levels of LMNB2 mRNA and LMNB2 protein by RT-PCR and western blotting (Fig. 4a, b).
Loss-of-function studies by siRNAs. a, b Expression levels of LMNB2 mRNA and LMNB2 protein 48 h after transfection with si-LMNB2 into LUAD cell lines. c Cell proliferation was evaluated by XTT assays. d Cell migration was determined by wound healing assay. e Cell invasion was determined by Matrigel invasion assay. These assays showed that inhibition of proliferation, migration, and invasion were observed in si-LMNB2-transfected cell lines (p < 0.0001). *p < 0.0001
Functional assays showed that cell proliferation, migration and invasive abilities were strongly inhibited by si-LMNB2 transfection in LUAD cells in comparison with mock- or miR-control-transfectants (Fig. 4c–e).
Expression of LMNB2 in LUAD clinical specimens
LMNB2 was mainly detected around the nuclear membrane. Our results showed that the LMNB2 expression levels of cancer tissues were higher than those of non-cancerous tissues (Fig. 5a–c).
Immunohistochemical staining of LMNB2 protein in LUAD specimens. Immunostaining of the LUAD specimens respectively collected from three patients who underwent lung surgery at Kagoshima University Hospital. a Lung adenocarcinoma with acinar growth pattern. b Lung adenocarcinoma with papillary and micropapillary growth pattern. c Lung adenocarcinoma with lepidic growth pattern. In all cases, high expression of LMNB2 was confirmed in the nuclear membrane of cancer cells
Discussion
A large number of studies have shown that expression of miR-145-5p is deeply involved in human physiological and pathological conditions, e.g., the cell cycle, differentiation, migration, invasion and apoptosis [26, 27]. In human embryonic stem cells (ESCs), expression of miR-145-5p promotes the differentiation of ESCs by suppressing OCT4, SOX2, and KLF4, thereby silencing the self-renewal program [28]. In the field of cancer, many studies have demonstrated that miR-145-5p is frequently downregulated in a wide range of diseases and acts as an anti-tumor regulator through targeting several oncogenes [29,30,31]. Importantly, the promoter region of pre-miR-145 has a p53 binding region. Indeed, its transcriptional activity is controlled by the pivotal tumor suppressor p53 [32].
It has been generally believed that the passenger strands of miRNAs are degraded and therefore have no function [33,34,35]. Thus, it was rather surprising that our analyses of miRNA expression signatures by RNA sequencing showed that some passenger strands were dysregulated in cancer tissues [9, 10]. Our functional assays showed that both strands of the miR-145 duplex and the miR-150 duplex act as anti-tumor miRNAs in several cancers [9, 14, 17, 18, 23]. We hypothesized that genes in which both strands of miRNAs were coordinately regulated were important in cancer cells. Based on this hypothesis, we showed that MTDH was regulated by both miR-145-5p and miR-145-3p in lung squamous cell carcinoma. Moreover, UHRF1 was modulated by both strands in bladder cancer [17, 18]. In addition, SPOCK1 was coordinately regulated by miR-150-5p and miR-150-3p in esophageal cancer and in head and neck cancer [9, 23]. These oncogenes (MTDH, UHRF1, and SPOCK1) were overexpressed in cancer tissues and their expression was significantly associated with poor prognosis of the patients [17, 18, 23].
In this study, we identified 4 genes (LMNB2, NLN, SIX4, and DDC) that were coordinately regulated by miR-145-5p and miR-145-3p in LUAD cells. Analysis of these genes should provide important information regarding the molecular pathogenesis of LUAD. In human genome, the miR-143 and miR-145 formed clustered miRNA in chromosome 5q32 region. Several studies showed that the miR-143/miR-145 clustered miRNAs were frequently downregulated in several types of cancers and acted as anti-tumor miRNAs [36, 37]. Our previous studies showed that miR-143-3p and miR-145-5p were coordinately regulated oncogenic Golgi membrane protein 1 (GOLM1) and hexokinase-2 (HK2) in prostate cancer and renal cell carcinoma, respectively [36, 37]. Interestingly, our present data showed that LMNB2 was directly regulated by miR-143-3p, miR-145-5p, and miR-145-3p in LUAD cells. Moreover, overexpression of LMNB2 was significantly associated with the poor prognosis of LUAD patients. For these reasons, we focused on the LMNB2 and further analyses were performed in this study.
Lamins are pivotal components of the nuclear lamina, and their interactions with chromatin and its binding partner act to regulate cell proliferation and differentiation [38]. Lamins are subdivided into A-types and B-types based on their biochemical properties [38]. In the human genome, lamin proteins are coded by three genes (LMNA, LMNB1, and LMNB2) [38]. Previous studies showed that aberrantly expressed lamin family members are observed in a wide range of cancers [39,40,41,42,43]. Data indicated that ectopic expression of anti-tumor miR-193a-3p suppressed expression levels of HNRNRU, G3BP1, UHRF1, and LMNB2 in the highly metastatic human lung cancer cell line SPC-A-1sci [44]. More recently, knockdown of LMNB2 in A549 and H1299 cells inhibited cell proliferation and increased apoptosis, indicating that aberrantly expressed LMNB2 enhanced cancer cell aggressiveness [45]. This study also showed that minichromosome maintenance complex component 7 (MCM7) was a binding partner of LMNB2 [45]. Interestingly, overexpression of the MCM family was reported in several cancers and their expression was involved in cancer pathogenesis [46,47,48]. TCGA database showed that high expression of MCM7 was significantly associated with poor prognosis of LUAD patients (p = 0.0083). Identification of the proteins with which LMNB2 interacts will lead to the elucidation of the molecular pathogenesis of LUAD.
In conclusion, downregulation of both strands of the miR-145 duplex (miR-145-5p and miR-145-3p) was detected in LUAD clinical specimens. Data showed that both strands possessed tumor suppressive abilities in LUAD cells. To the best of our knowledge, this is the first report showing that anti-tumor miR-145-5p and miR-145-3p coordinately regulated LMNB2 in LUAD cells. Overexpression of LMNB2 enhanced cancer cell aggressiveness and its expression was involved in LUAD pathogenesis. Further studies of anti-tumor miRNA-modulated pathways in LUAD may provide novel insights into the molecular pathogenesis of this disease.
References
Global Burden of Disease Cancer Consortium. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–48.
Travis WD. Pathology of lung cancer. Clin Chest Med. 2011;32:669–92.
Arai T, Kuroishi T, Saito Y, Kurita Y, Naruke T, Kaneko M. Tumor doubling time and prognosis in lung cancer patients: evaluation from chest films and clinical follow-up study. Jpn J Clin Oncol. 1994;24:199–204.
Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J Transl Med. 2016;14:143.
Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004.
Ramassone A, Pagotto S, Veronese A, Visone R. Epigenetics and microRNAs in cancer. Int J Mol Sci. 2018;19:E459.
Koshizuka K, Nohata N, Hanazawa T, Kikkawa N, Arai T, Okato A, et al. Deep sequencing-based microRNA expression signatures in head and neck squamous cell carcinoma: dual strands of pre-miR-150 as antitumor miRNAs. Oncotarget. 2017;8:30288–304.
Mizuno K, Mataki H, Arai T, Okato A, Kamikawaji K, Kumamoto T, et al. The microRNA expression signature of small cell lung cancer: tumor suppressors of miR-27a-5p and miR-34b-3p and their targeted oncogenes. J Hum Genet. 2017;62:671–8.
Arai T, Okato A, Yamada Y, Sugawara S, Kurozumi A, Kojima S, et al. Regulation of NCAPG by miR-99a-3p (passenger strand) inhibits cancer cell aggressiveness and is involved in CRPC. Cancer Med. 2018;7:1988–2002.
Goto Y, Kurozumi A, Arai T, Nohata N, Kojima S, Okato A, et al. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br J Cancer. 2017;117:409–20.
Koshizuka K, Hanazawa T, Kikkawa N, Arai T, Okato A, Kurozumi A, et al. Regulation of ITGA3 by the anti-tumor miR-199 family inhibits cancer cell migration and invasion in head and neck cancer. Cancer Sci. 2017;108:1681–92.
Okato A, Arai T, Kojima S, Koshizuka K, Osako Y, Idichi T, et al. Dual strands of pre-miR150 (miR1505p and miR1503p) act as antitumor miRNAs targeting SPOCK1 in naive and castration-resistant prostate cancer. Int J Oncol. 2017;51:245–56.
Yamada Y, Koshizuka K, Hanazawa T, Kikkawa N, Okato A, Idichi T, et al. Passenger strand of miR-145-3p acts as a tumor-suppressor by targeting MYO1B in head and neck squamous cell carcinoma. Int J Oncol. 2018;52:166–78.
Yonemori K, Seki N, Idichi T, Kurahara H, Osako Y, Koshizuka K, et al. The microRNA expression signature of pancreatic ductal adenocarcinoma by RNA sequencing: anti-tumour functions of the microRNA-216 cluster. Oncotarget. 2017;8:70097–115.
Mataki H, Seki N, Mizuno K, Nohata N, Kamikawaji K, Kumamoto T, et al. Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget. 2016;7:72084–98.
Matsushita R, Yoshino H, Enokida H, Goto Y, Miyamoto K, Yonemori M, et al. Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell aggressiveness. Oncotarget. 2016;7:28460–87.
Kamikawaji K, Seki N, Watanabe M, Mataki H, Kumamoto T, Takagi K, et al. Regulation of LOXL2 and SERPINH1 by antitumor microRNA-29a in lung cancer with idiopathic pulmonary fibrosis. J Hum Genet. 2016;61:985–93.
Suetsugu T, Koshizuka K, Seki N, Mizuno K, Okato A, Arai T, et al. Downregulation of matrix metalloproteinase 14 by the antitumor miRNA, miR-150-5p, inhibits the aggressiveness of lung squamous cell carcinoma cells. Int J Oncol. 2018;52:913–24.
Mataki H, Enokida H, Chiyomaru T, Mizuno K, Matsushita R, Goto Y, et al. Downregulation of the microRNA-1/133a cluster enhances cancer cell migration and invasion in lung-squamous cell carcinoma via regulation of Coronin1C. J Hum Genet. 2015;60:53–61.
Mizuno K, Seki N, Mataki H, Matsushita R, Kamikawaji K, Kumamoto T, et al. Tumor-suppressive microRNA-29 family inhibits cancer cell migration and invasion directly targeting LOXL2 in lung squamous cell carcinoma. Int J Oncol. 2016;48:450–60.
Osako Y, Seki N, Koshizuka K, Okato A, Idichi T, Arai T, et al. Regulation of SPOCK1 by dual strands of pre-miR-150 inhibit cancer cell migration and invasion in esophageal squamous cell carcinoma. J Hum Genet. 2017;62:935–44.
Kumamoto T, Seki N, Mataki H, Mizuno K, Kamikawaji K, Samukawa T, et al. Regulation of TPD52 by antitumor microRNA-218 suppresses cancer cell migration and invasion in lung squamous cell carcinoma. Int J Oncol. 2016;49:1870–80.
Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci. 2016;2:e67.
Kent OA, McCall MN, Cornish TC, Halushka MK. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res. 2014;42:7528–38.
Morgado AL, Rodrigues CM, Sola S. MicroRNA-145 regulates neural stem cell differentiation through the Sox2-Lin28/let-7 signaling pathway. Stem Cells. 2016;34:1386–95.
Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–58.
Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, et al. miR-145, miR-133a and miR-133b: tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:2804–14.
Liu SY, Li XY, Chen WQ, Hu H, Luo B, Shi YX, et al. Demethylation of the MIR145 promoter suppresses migration and invasion in breast cancer. Oncotarget. 2017;8:61731–41.
Mei LL, Wang WJ, Qiu YT, Xie XF, Bai J, Shi ZZ. miR-145-5p suppresses tumor cell migration, invasion and epithelial to mesenchymal transition by regulating the Sp1/NF-kappaB signaling pathway in esophageal squamous cell carcinoma. Int J Mol Sci. 2017;18:E1833.
Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106:3207–12.
Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4.
Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–60.
Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–20.
Kojima S, Enokida H, Yoshino H, Itesako T, Chiyomaru T, Kinoshita T, et al. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J Hum Genet. 2014;59:78–87.
Yoshino H, Enokida H, Itesako T, Kojima S, Kinoshita T, Tatarano S, et al. Tumor-suppressive microRNA-143/145 cluster targets hexokinase-2 in renal cell carcinoma. Cancer Sci. 2013;104:1567–74.
Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar DZ, Barkan R, et al. Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med. 2009;13:1059–85.
Broers JL, Ramaekers FC. The role of the nuclear lamina in cancer and apoptosis. Adv Exp Med Biol. 2014;773:27–48.
Butin-Israeli V, Adam SA, Goldman AE, Goldman RD. Nuclear lamin functions and disease. Trends Genet. 2012;28:464–71.
Irianto J, Pfeifer CR, Ivanovska IL, Swift J, Discher DE. Nuclear lamins in cancer. Cell Mol Bioeng. 2016;9:258–67.
Kong L, Schafer G, Bu H, Zhang Y, Zhang Y, Klocker H. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis. 2012;33:751–9.
Sakthivel KM, Sehgal P. A novel role of lamins from genetic disease to cancer biomarkers. Oncol Rev. 2016;10:309.
Deng W, Yan M, Yu T, Ge H, Lin H, Li J, et al. Quantitative proteomic analysis of the metastasis-inhibitory mechanism of miR-193a-3p in non-small cell lung cancer. Cell Physiol Biochem. 2015;35:1677–88.
Ma Y, Fei L, Zhang M, Zhang W, Liu X, Wang C, et al. Lamin B2 binding to minichromosome maintenance complex component 7 promotes non-small cell lung carcinogenesis. Oncotarget. 2017;8:104813–30.
Liu YZ, Wang BS, Jiang YY, Cao J, Hao JJ, Zhang Y, et al. MCMs expression in lung cancer: implication of prognostic significance. J Cancer. 2017;8:3641–7.
Nishihara K, Shomori K, Fujioka S, Tokuyasu N, Inaba A, Osaki M, et al. Minichromosome maintenance protein 7 in colorectal cancer: implication of prognostic significance. Int J Oncol. 2008;33:245–51.
Simon NE, Schwacha A. The Mcm2-7 replicative helicase: a promising chemotherapeutic target. Biomed Res Int. 2014;2014:549719.
Acknowledgements
The present study was supported by KAKENHI grants 17K09660 and 18K09338.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Misono, S., Seki, N., Mizuno, K. et al. Dual strands of the miR-145 duplex (miR-145-5p and miR-145-3p) regulate oncogenes in lung adenocarcinoma pathogenesis. J Hum Genet 63, 1015–1028 (2018). https://doi.org/10.1038/s10038-018-0497-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-018-0497-9
This article is cited by
-
Expression and gene regulatory network of SNHG1 in hepatocellular carcinoma
BMC Medical Genomics (2021)
-
Molecular pathogenesis of breast cancer: impact of miR-99a-5p and miR-99a-3p regulation on oncogenic genes
Journal of Human Genetics (2021)
-
Regulation of aberrantly expressed SERPINH1 by antitumor miR-148a-5p inhibits cancer cell aggressiveness in gastric cancer
Journal of Human Genetics (2020)
-
RNA sequencing-based microRNA expression signature in esophageal squamous cell carcinoma: oncogenic targets by antitumor miR-143-5p and miR-143-3p regulation
Journal of Human Genetics (2020)
-
Pan-cancer analysis reveals cooperativity of both strands of microRNA that regulate tumorigenesis and patient survival
Nature Communications (2020)