Abstract
Lymphoblastoid cell lines (LCLs) have been by far the most prevalent cell type used to study the genetics underlying normal and disease-relevant human phenotypic variation, across personal to epidemiological scales. In contrast, only few studies have explored the use of LCLs in functional genomics and mechanistic studies. Two major reasons are technical, as (1) interrogating the sub-cellular spatial information of LCLs is challenged by their non-adherent nature, and (2) LCLs are refractory to gene transfection. Methodological details relating to techniques that overcome these limitations are scarce, largely inadequate (without additional knowledge and expertise), and optimisation has never been described. Here we compare, optimise, and convey such methods in-depth. We provide a robust method to adhere LCLs to coverslips, which maintained cellular integrity, morphology, and permitted visualisation of sub-cellular structures and protein localisation. Next, we developed the use of lentiviral-based gene delivery to LCLs. Through empirical and combinatorial testing of multiple transduction conditions, we improved transduction efficiency from 3% up to 48%. Furthermore, we established strategies to purify transduced cells, to achieve sustainable cultures containing >85% transduced cells. Collectively, our methodologies provide a vital resource that enables the use of LCLs in functional cell and molecular biology experiments. Potential applications include the characterisation of genetic variants of unknown significance, the interrogation of cellular disease pathways and mechanisms, and high-throughput discovery of genetic modifiers of disease states among others.
Similar content being viewed by others
Introduction
The investigation of human traits and genetic disease has greatly benefited from the collections of human derived non-cancerous cell lines. Among the different types that have been used, the generation and use of lymphoblastoid cell lines (LCLs) from individual human blood samples has been the most prevalent by far. The procedure for generating LCL lines is easy and has remained relatively unchanged for over two decades [1, 2]. Starting from a small sample of human blood, lymphocytes are isolated, and exposed to Epstein–Barr Virus (EBV), which infects predominantly B lymphocytes. The removal of T lymphocytes (via ongoing culture or actively via treatments e.g., with Cyclosporin A) then results in pure populations of transformed B cells, termed LCLs. In these cells, the EBV establishes a latent infection, where it remains predominantly episomal (10–50 copies per cell), and expresses low levels of viral genes (e.g., EBNA1, −2, 3 A, 3B, LP and LMP1, −2A, and 2B) that confer immortalisation [3]. The simple ease of production, maintenance, and cryogenic storage of LCLs, together with the low (>0.03%) rate of somatic mutation [4] has seen LCLs be used as a standard and renewable resource of an individual’s specific biomolecules, DNA, RNA, and protein [3,4,5]. Tens or even hundreds of thousands of individual cell lines have been banked internationally, from a variety of ethnic backgrounds and disease states [6,7,8]. LCLs have been used for an enormous multitude of genotype–phenotype studies involving small to very large human cohorts, spanning disease-related gene discovery, genome-wide association, and pharmacogenomic studies, among many others [9,10,11,12,13,14,15,16,17,18,19]. Whereas the use of isolated biomolecules from these cell lines has been of great advantage, few studies have ever explored the use of LCLs in more in-depth cell biology-based experiments, despite many potential benefits, e.g., using patient-derived material to understand the impact of genetic mutation on gene function, to study the molecular and cellular mechanisms of disease, and to develop personalised medicines. The lack of use of LCLs in cell biology assays stems from two inherent features of LCLs (1) they are non-adherent cells making sub-cellular localisation techniques such as immunofluorescence technically difficult, and (2) they are refractory to standard genetic manipulation techniques such as transfection. These challenges can however can be overcome, and a handful of studies (focusing on EBV infection itself and/or immunology), have presented such data [20,21,22,23,24,25,26,27,28,29,30,31,32,33]. Although encouraging, the methodological and experimental details related to the techniques in these studies varied greatly, and are in all cases insufficiently described to readily reproduce without additional expertise and knowledge. Furthermore, there has been no systematic comparisons of experimental parameters that enable derivation of optimised approaches. As a consequence, studies interrogating the effect of genetic variation on cellular and molecular processes in LCLs has remained extremely limited, even if by simple comparison with the > 10,000 current studies that have employed their use.
Stemming from our motivation to conduct functional genomic studies in LCLs, and the paucity of methodological approaches in the literature that restricted us, in this study we developed and empirically tested imaging and gene delivery methods. First, we develop a method that facilitates adhesion of LCLs to coverslips via use of cell-adhesive polymers, which enabled sub-cellular protein localisation studies via standard immunofluorescence techniques. Next, we developed lentiviral based cell transduction technologies that allowed us to generate near pure populations of genetically manipulated LCLs that can be maintained over several passages. Our relatively simple methods (supplied also as detailed laboratory based protocols) employ commercially available resources, and thus unlock avenues for the use of LCLs in cell biology, and as such facilitate further understanding of the effects of genetic change, whether it be disease related or part of natural human variation.
Materials and methods
Please see Online Resource 1 for detailed laboratory protocols of our optimised methods.
LCLs culture
Low passage LCLs obtained from healthy individuals were grown as previously described [34]. In brief, cells were maintained in LCL growth media (Rosewell Park Memorial Institute media supplemented with 10% fetal calf serum (FCS), 2mM l-glutamine and 1% penicillin–streptomycin, Sigma). Cells were refed every third day and passaged 1:3 once per week. For transduction, 1 × 105 LCLs were plated in 0.5 ml in a well of a 24-well plate in growth media supplemented with polybrene (Sigma) and viral particles as described in main text. For centrifugation-assisted transduction, cells and virus and polybrene were mixed in 1 ml of growth media in a centrifuge tube and subjected to centrifugation at 2000 × g. Cells were resuspended and plated in 24-well plate as described above. Morning following overnight transduction, cells were collected and centrifuged and virus-containing supernatant removed. Cells were then resuspended in 4 ml of LCL growth media and cultured in an upright T25 flask. Media was changed every 3–4 days. Where noted, blasticidin (Thermofisher) was added to media at given concentrations. Trypan blue was used for viable cell counts, with at least 500 cells scored for each replicate.
Immunostaining
Glass coverslips were acid washed (1 N HCl) at 60 °C overnight, extensively washed in reverse osmosis water and sterilised using 100% ethanol. Coverslips were coated for 4 h using a solution of 1 mg/ml poly-l-lysine or 0.1 mg/ml poly-l-ornithine as per manufactures instruction (Sigma). Coverslips were then washed and sterilised as described above. Pre-coated coverslips are commercially available (e.g., Corning BioCoat). Coverslips were placed in 24-well plate. LCLs were centrifuged at 320 × g for 5 min. Supernatant was discarded and the cells were resuspended in phosphate-buffered saline (PBS) and triturated into single cell suspension. A total of 4 × 105 cells were added to coverslips in a 500 µl volume. Cells were allowed to settle and adhere for 5 or 10 min. In total, 125 μl (25% of the cell suspension volume) of 4% paraformaldehyde (PFA) was then added in order to spike the cell suspension and minimise potential osmotic stress caused by the PFA. Following a 2-min incubation at room temperature (RT), the solution was removed and replaced with fresh 4% PFA. Cells were fixed for 30 min at RT without agitation. Cells were washed gently using PBS three times before subjected to standard immunofluorescent staining (e.g., as previously described [35]). Care was taken during wash steps not to completely remove all wash solutions (to avoid physical dislodgement of cells by solution meniscus), complimented by the slow administering of excessive volumes of wash solution (3 ml/well/wash). Primary antibodies: THOC2 (303–630 A; Bethyl Laboratories), GOSR2 (mouse pAB, Abnova). Alexaflor conjugated secondary antibodies were from Thermo Fisher. 4′,6-diamidino-2-phenylindole (DAPI) and Phalloidin were used as per manufactures instructions (Thermofisher).
Lentivirus production
The lentiviral system employed has been extensively described ([36, 37] and www.LentiGO-Vectors.de). Lentiviral-packaging plasmids (pMDLg/pRRE, pRSV-Rev, and pMD2.0 G) and transfer vector (pLego-G/BSD) were obtained from Addgene and prepared using endo-toxin free DNA kits (Qiagen). Lentiviral particles were generated by transfecting plasmids into HEK293T broad cells. HEK293T cells were grown in Dulbecco's Modified Eagle Medium supplemented with 10% FCS, 2 mM l-Glutamine and 1% penicillin–streptomycin (all from Sigma). A total of 1 × 107 cells were plated in a T75 culture flask. The following day, cells were transfected by Lipofectamine 2000 reagent (Thermo Fisher Scientific) using manufactures instructions: 15 µg transfer vector, 10 µg pMDLg/pRRE, 5 µg pRSV-Rev, and 2 µg pMD2.0 G. The medium was replaced the next morning. Viral supernatants were collected 48 h later, passed through a 0.45 µm filter, and pelleted using ultracentrifugation 20,000 rpm for 90 min at 4 °C. The pellet was resuspended in PBS to gain a 100 × concentrated stock, which was aliquoted and frozen at − 80 °C. A frozen aliquot was thawed and used to determine viral titre as previously described [35]. Concentrated titre was ~1 × 109 infective units per ml.
FACS
For fluorescence-activated cell sorting (FACS) profiling, cells were collected and washed twice in PBS before being triturated into single cells and resuspended in fixative (2% formaldehyde, 2% glucose and 0.02% sodium azide in PBS). FACS profiling was conducted using Becton Dickinson LSR Fortessa instrument. Non-transduced cultures were used to set negative and size gates. For cell sorting, cells were triturated into single cells and resuspended in FACS sorting buffer (Ca/Mg++ free PBS with 1 mM ethylenediaminetetraacetic acid, 25 mM HEPES pH 7.0 and 1% FCS). Cells were sorted using the Beckman Coulter MoFlo® Astrios instrument.
Microscopy
Fluorescence was viewed using the inverted Axiovert microscope (Carl Zeiss, Jena, Germany) fitted with an HBO 120 lamp (Carl Zeiss). Images were captured using an Axiocam Mrm camera and Axio Vs40 v4.5.0.0 software (Axiovision, Carl Zeiss).
Statistical analysis
All experiments conducted in biological triplicate (individual LCL cell lines). Data points on graphs represent the average of biological triplicate, error bars represent ± standard deviation. Two tailed Student’s t test was applied to interrogate significance, *p < 0.05.
Results
Sub-cellular imaging in LCLs
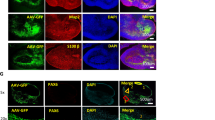
The use of LCLs in cell biology assays requires the ability to interrogate intracellular spatial information of single cells. The methodology to conduct such intracellular approaches, such as fluorescent staining of organelles or immunohistological detection of protein localisation, are rarely utilised in LCLs owing to the technical difficulties associated with their non-adherent nature. That the cultures are floating, aggregates of cells with weak cell–cell adhesion greatly complicates the staining process and subsequent mounting for microscopic imaging. We thus sought to develop a methodology to overcome these issues. Our aim was to adhere dissociated LCLs to coverslips using gentle techniques that balanced the needs of (i) maintaining cellular integrity and morphology, and (ii) preventing loss of cells that occur by washing procedures in standard staining protocols. We initially explored the cytospin technique [38] used commonly to conduct gross staining of blood-derived cells. A single cell suspension of LCLs was pipetted onto microscope slides and centrifuged followed by desiccation. We found that although cells were adhered, the integrity and morphology of the LCLs was compromised (data not shown). We took an alternative approach by attempting to adhere cells to coverslips, which we coated with cell matrix substrates, poly-l-lysine or poly-l-ornithine. Both of these substrates impart positive charge to the coverslip surface that promotes cell adhesion. Preparations of dissociated LCLs were applied to the coverslips for either 5 or 10 minutes to allow adhesion prior to fixation. Cells were stained with DAPI and phalloidin to reveal the nuclear and actin cytoskeletal structures, respectively, as a means to assess cell morphology (Fig. 1a). The combination of poly-l-lysine and 5 minutes adherence provided significant benefits in maintaining the typical rounded morphology of LCLs (81% of cells), compared with longer adherence times and/or use of poly-l-ornithine (Fig. 1b). Using this optimised approach, we next tested if the process was conducive to subsequent immunofluorescent staining involving several wash steps. We were able to faithfully identify the nuclear and Golgi compartments of LCLs via immunostaining of nuclear localised mRNA export factor THOC2 [39], and the Golgi localised protein GOSR2 [40] (Fig. 1c). Together these data identify robust methods to interrogate sub-cellular compartments and protein localisation in LCLs (for details, see Online Resource 1 for detailed laboratory based protocols).
LCLs attached to poly-l-lysine-coated coverslips maintain morphology and are conducive to immunofluorescent staining techniques. a, b LCLs were dissociated and seeded onto coverslips coated with either poly-l-lysine or poly-l-ornithine and allowed to attach for either 5 or 10 mins before paraformaldehyde fixation. a Cells were co-stained with DAPI and phalloidin to reveal nuclei and overall morphology. Closed arrow heads indicate examples of cells with typical rounded LCLs morphology; open arrow heads indicate cells with abnormal morphologies. b Quantification of cells displaying typical rounded morphology. Error bars represent standard deviation. *significantly different to poly-l-lysine: 5 min condition, p < 0.01 by Student’s two-tailed t-test. c Cells were stained as above following immunofluorescent staining of both a nuclear protein (THOC2) and a Golgi protein (GOSR2). Top panel represents low magnification and bottom panel represents high magnification representative images
Lentiviral transduction of LCLs
Next, we sought methods to manipulate gene expression in LCLs. Previously, we had tested and attempted to optimise numerous DNA transfection conditions for use in LCLs. Using a variety of methods and conditions, including lipid based deliveries (Lipofectamine 2000, Lipofectamine LTX (Thermo Fisher Scientific), FuGene (Promega)), and electroporation-based deliveries (Gene pulse, BioRad [41] and Nucleofection 2B, B-cell kit, Lonza [33]), we were unable to achieve > 3% transfection efficiency and observed high levels of cell death (data not shown). To overcome these issues, we compared lentiviral transduction as a means to deliver ectopic genetic material [20]. We employed the well-characterised Lentiviral Gene Ontology (LeGO) series of lentiviral transfer vectors, as they are freely available from Addgene and have versatility to co-express selection cassettes (various antibiotic resistance genes or fluorescent reporters or fusions of both) in cis with cassettes enabling gene knock-down (via shRNA), complementary DNA expression or both [36, 37, 42, 43]. To test our methods, we employed a LeGO transfer vector that expresses an Enhanced Green Fluorescent Protein: blasticidin resistance fusion protein (EGFP:BSDR), thus enabling profiling of transduction rates using fluorescent microscopy or FACS profiling, and a means to select for transduced cells using either blasticidin or FACS (Fig. 2a). We produced high titre viral preparations (~ 109 infective units/ml) using lipid based transfection of HEK293T cells and subsequent ultracentrifugation of viral supernatants. We used aliquots of the same batch of the virus for all subsequent experiments. In our experiments we used overnight transduction and assayed the cells at least 1 week later (up to 4 weeks later in some cases) to gain an accurate measure of expression from genomic integrated transgenes, as opposed to transient episomal expression (see timeline, Online Resource 1). Initially, we tested the effects of increasing multiplicity of infection (MOI) and the addition of polybrene, a carrier protein known to aid docking of lentiviral particles to the cell membrane. The baseline transduction conducted at an MOI of 25 resulted in transduction of only 2.8% of cells (as assayed using EGFP:BSDR expression and FACS profiling; Fig. 2c). The percentage of transduced cells increased linearly when additional virus was added, to a maximum of 5.7% at MOI 100. The addition of polybrene at 4 µg/ml increased the transduction rates at all MOIs tested to a maximum of 9.3% (seen for both MOI 50 and 100), and addition of 8 µg/ml further improved this outcome (maximum of 12.5% of cells transduced). Interestingly, this maximum rate was observed at all MOIs (25–100). These data suggest that polybrene had the highest effect when the number of viral particles was limiting. Further increases of polybrene concentrations were associated with loss of cell viability (data not shown). We used 8 µg/ml of polybrene in all subsequent experiments. Whereas increases in MOI generally increase transduction rates of various cell types, an upper limit exists beyond which cell viability is affected. To determine this limit, we tested the proliferation of LCLs following overnight transduction of lentiviral particles at increasing MOIs. There was no significant change in the proliferation rates of non-transduced LCLs compared to LCLs transduced at MOIs of 25 and 50 (Fig. 3a). At MOI 100, a slight and significant 21% reduction in proliferation was observed, whereas at the highest MOIs tested (200 and 400), a ~ 52% reduction in cell proliferation was observed. Thus, for the subsequent studies we focused on MOI 25–100. Although there is a fourfold difference in the amount of virus in this range, the percentage of transduced cells (i.e., expressing EGFP:BSDR) was ~ 9.1% for all conditions.
Polybrene enhances the efficiency of lentiviral transduction of LCLs. a Schematic of the transgene delivered by lentivirus, employing the spleen focus forming virus (SFFV) promoter to drive the expression of the fusion gene Enhanced Green Fluorescent Protein: blasticidin Resistance (EGFP:BSDR). b, c LCLs were transduced with lentiviral particles encoding EGFP:BSDR at multiplicity of infection of 25, 50, and 100 in presence of 0, 4, or 8 µg/ml of polybrene. b Representative images of LCL colonies expressing EGFP at MOI 25. c Percentage of cells expressing EGFP:BSDR as assayed by FACS. Note that further increases in polybrene concentration impacted on cell viability (not shown). Graphed results represent the mean of three independent experiments. Error bars represent standard deviation. *significantly different to 0 µg/ml conditions, p < 0.05 by Student’s two-tailed t test
Improved methods for LCLs transduction using lentiviral particles. LCLs were transduced with lentiviral particles encoding the EGFP:BSDR fusion protein. a–c High multiplicity of infection (MOI) does not improve transduction rates. a Cell viability counts resulting from variable MOI ratios. Note significant impact on cell viability above MOI 100. b Representative image of LCL colonies expressing EGFP:BSDR at MOI 25. c Percentage of cells expressing EGFP:BSDR as assayed by FACS profiling of cultures transduced at ratios of MOI 25, 50, and 100. Note increased MOI does not improve transduction rates. d–f Centrifugation during transduction improves transduction rate. Cells and viral particles were centrifuged briefly (1 h at 2000 × g) before culture. d Representative images of LCL colonies expressing EGFP:BSDR. e FACS profiles of transduced cultures. f Quantification of transduced cells assayed using FACS. g–i Sequential round of LCLs transduction improves transduction efficiency. LCLs were subjected transduction at two sequential passages. g Representative images of LCL colonies expressing EGFP:BSDR. h FACS profiles of transduced cultures. i Quantification of transduced cells assayed using FACS. Graphed results represent the mean of three independent experiments. Error bars represent standard deviation. *p < 0.05 by Student’s two-tailed t-test
To seek methods that would improve transduction efficiency we assayed both the effects of a centrifugation step and the outcome of sequential rounds of transduction. Centrifugation has been proposed to increase transduction rates of some cell types by enhancing the interaction of viral particles with cells. We added virus to LCLs at MOI of 25–100 and immediately centrifuged the cell/virus mixture at 2000 × g for 1 h. Cells were resuspended and cultured for an additional week before being assayed by FACS. This method resulted in significant increases in cell transduction across all MOIs tested (29.7% increase at MOI 25 up to maximum 62.7% increase at MOI 100, giving a transduction rate of 14.9%; Fig. 3f). Next, we investigated the additive effect of a sequential round of transduction by following an initial round of transduction at MOIs 25 and 50 with a subsequent round 1 week later, and assaying after a further week of growth. At MOI 25 and 50, the sequential approach increased transduction rates by 2.1- and 2.5-fold respectively, resulting in overall transduction rate of 37.8–40.1% (Fig. 3i). Together, these data show that through strategic alterations to the transduction protocol, we were able to increase the initial transduction of LCLs from 2.8% (MOI 25 without polybrene), up to 11.9% via addition of polybrene, and subsequently up to 12.2–14.4% via centrifugation, and to a maximum of 37.8–40.1% following sequential transduction at MOIs of 25 and 50 (for details, see Online Resource 1 for detailed laboratory based protocols).
Selection of transduced LCLs
Although transduction rates of 40% may be sufficient for some experimental applications, we sought methods to further select for transduced cells with aim of producing a near homogenous population of transduced LCLs. Initially, we exploited the expression of a blasticidin resistance fusion gene in our lentiviral transfer vector. We first tested the susceptibility of LCLs to blasticidin by treating non-transduced cells with varying concentrations and across different lengths of exposure time. Even treatment with the lowest amount of blasticidin (5 µg/ml) had significant effect on cell viability (73.8% cell death after 4 days, and 92.0% cell death after 8 days; Online Resource 1). Concentrations of between 10 and 80 µg/ml, further enhanced cell death. As the differences between 10 and 40 µg/ml were not significant, we selected 10 µg/ml for further experiments, which after 4 days treatment resulted in 89.6% cell death. Next we transduced cells (MOI 25 with 8 µg/ml polybrene) and after two weeks of further culture to expand cell number, we treated cultures with or without blasticidin for 4 days. Prominent cell death was observed in first days of blasticidin treatment as expected, with resistant cells subsequently expanding. Cells were grown for a further two weeks to allow recovery. Consistent with previous results, non-treated cultures contained 9.5% cells expressing EGFP:BSDR (for details, see Online Resource 1). The treatment with blasticidin improved the percentage of EGFP:BSDR expressing cells by 2.8-fold, reaching a total of 26.75% of transduced cells. Thus, although blasticidin improved the purity of transduced cells in the cultures by 2.8-fold, it did not achieve pure populations of EGFP-expressing cells. We reasoned that if we were able to increase the initial transduction rate from 9.5% to a higher rate (e.g. ~ 40%) then a 2.8-fold enhancement via blasticidin treatment should result in near homogeneity (e.g., 100%) of EGFP:BSDR expressing cells.
We also sought to compare blasticidin selection with FACS of EGFP:BSDR-expressing cells as a means of transduced cell purification. To increase the initial transduction rate of cells, we combined the methods of polybrene (8 µg/ml), centrifugation (2000 × g for 1 h), and sequential transduction (2 transductions separated by 1 week). We next either continued to culture the cells, or subjected them to either 1 week of blasticidin selection or FACS to isolate EGFP-expressing cells. All cells were grown for an additional 1–2 weeks (1 week for blasticidin, 2 weeks for FACS selections, such that all cells were grown for 4 weeks total since initial transduction) and then FACS profiled for EGFP:BSDR expression. Our combined method of transduction indeed increased the transduction rate of cells to 47.2% (Fig. 4c). Blasticidin selection significantly improved the purity of these cultures, however, only by 1.3-fold, resulting in a culture with 62.1% EGFP:BSDR expressing cells. In comparison, FACS of cells further enhanced the percentage of EGFP:BSDR-expressing cells in the culture, achieving a total of 85.9%. Furthermore, as evidenced by the FACS profile, cells purified by FACS-contained cells expressing much higher levels of EGFP:BSDR, suggesting this method of purification has the ability to isolate cells expressing the highest amounts of integrated genetic material delivered by transduction (Fig. 4b). Together, this work outlines methods to greatly enhance the initial transduction rate (from 2.7 to 47.2%) and identifies FACS as the method of choice to further purify EGFP-expressing cells to achieve near homogeneous cultures of transduced cells with stable transgene expression (for details, see Online Resource 1 for detailed laboratory based protocols).
Efficient transduction and selection of transduced LCLs. LCLs were transduced with lentiviral particles encoding an EGFP:BSDR fusion protein at a MOI of 25 using a combination of both sequential (two rounds) and centrifugation methods. Transduced cells were then subjected to either blasticidin selection or FACS purification and cultured a further 2 weeks before analysis. a Representative images of LCL colonies expressing EGFP:BSDR. b FACS profiles of transduced cultures. c Quantification of transduced cells assayed using FACS. Graphed results represent the mean of 3 independent experiments. Error bars represent standard deviation. *p < 0.05 by Student’s two-tailed t test
Discussion
Biomedical research has greatly benefited from the availability and use of biological materials, and cell lines in particular, from individuals of different ancestry or health status. The generation and exploitation of human blood-derived LCL lines, for the advancement of the knowledge in genetics and genomics, in particular, has been transforming. Relative ease of access to the cell source, that is blood, coupled with ease of cell immortalisation, long-term culture, cryopreservation, and low somatic mutation rate has provided researchers with continuous supply of biomolecules matched against different human genotypes and phenotypes [5]. LCL lines have been bio-banked in huge numbers (e.g., Corriell Institute) and have been instrumental in integrating genomic, epigenomic, transcriptomic, proteomic, metabolomic, and pharmacogenomic studies, in both small and large cohorts, spanning personalised through to epidemiological studies [9, 10, 12,13,14,15,16,17,18,19, 28, 44]. LCLs have been intensively characterised. To name a few, LCLs have been crucial for the success of major projects like the International Haplotype Mapping (HapMap) project [6], Encyclopaedia of DNA Elements (ENCODE) projects [8], Genotype-Tissue Expression (GTEx) studies [15, 45], 1000 Genomes and Geuvadis Projects [7, 14]. LCLs facilitated unprecedented insights into the effects of normal and disease causing genetic variation. In contrast to their universal use as a basic source of biomolecules of an individual, only few studies have utilised LCLs in other more in-depth cell biology based investigations. Some have exploited the immunological (B cell) origin of LCLs and exposed them to antigens and/or cytokines in order to interrogate the cell and molecular mechanisms of immune responses [31]. Others have used genotoxic compounds to interrogate DNA damage response mechanisms in cell lines obtained from healthy individuals and patients with germ line cancerous mutations [9]. In such examples, cells are exposed en masse to bioactive treatments and responses are monitored. In-depth interrogations of genetic pathways typically require genetic manipulation to understand the cell and molecular mechanisms they affect. Approaches including gene silencing and overexpression are gold-standard techniques in this regard. In addition, spatial knowledge of how molecular mechanisms operate (e.g., signal transduction) within cells, require sub-cellular imaging of cellular proteins and organelles. Unfortunately, owing to the refractory nature of LCLs to DNA transfection and sub-cellular imaging techniques, the use of LCLs in cell biology and disease modelling has lagged far behind its potential. With a goal to perform function genomic studies in LCLs, we searched the literature for protocols that would facilitate sub-cellular imaging and gene manipulation of LCLs. Although finding several examples of each, the methodology varied greatly between labs, and was typically insufficiently described [20,21,22,23,24,25,26,27,28,29,30,31,32,33]. Relating to retroviral gene delivery, details such as multiplicity of infection, cell density, centrifugation conditions, use of carrier proteins, time of transduction, culture conditions (to name only some parameters) are all important for transduction efficiency and cell viability, but were rarely and incompletely referenced. Furthermore, transduction efficiencies were almost universally absent in these studies making it impossible to compare and adopt best practise. Likewise, immunofluorescent procedures were not readily transferable from most descriptions, and lack comparative analysis. As such, here we aimed to work up and describe in detail, simple and robust techniques using commercially available products to image and genetically manipulate LCLs.
LCLs grow as non-adherent cells in suspension, which is the main hurdle to their use in cell imaging. Although the cytospin method [38] has been described as a means to stick the cells to, e.g., the glass surface, the required equipment is not available in most labs, and (as in our experience) it can cause loss of structural (and hence spatial) integrity of the cell. We reasoned that other cell culture surface substrates would facilitate adherence of LCLs to coverslips. Although both poly-l-lysine and poly-l-ornithine facilitated adherence of LCLs to coverslips, the typically rounded LCL morphology was the most robust only when using poly-l-lysine. Adherence of cells took only 5 min, and after subsequent fixation of cells for 30 min with PFA, we were able to perform standard immunofluorescent staining protocols. Using this approach, LCLs remained attached and their cell morphology and integrity was not dramatically affected. We successfully detected sub-cellular protein localisation and organelles, supporting the utility of our method. We believe our method has widespread applications. For example, it will aid functional characterisation of disease associated genetic variants of unknown significance in many ways; from assaying the effect of coding DNA variants on protein stability or localisation, non-coding variants on gene expression and regulation, to the identification of cellular and molecular phenotypes associated with health or disease.
To manipulate gene expression in LCLs, we initially tested a variety of the DNA transfection techniques, including a range of lipid-based and electroporation methods [25, 33, 41], all without success (<3% transfection rates and high cell death). We thus assayed the utility of lentiviral-based gene delivery. Many lentiviral vector systems are freely available from Addgene, such as the LeGO vectors we employed. Similar other vector systems are commercially available. Furthermore, being a retrovirus, lentiviral delivery of transgenes results in stable genomic integration, providing long-term expression permitting creation of stable LCLs [20, 32]. Indeed, we observed stable expression of transgenes for over one month post transduction. Our data show that through empirical and combinatorial testing of multiple transduction conditions and criteria, we improved our initial transduction rate of 3–40%. Addition of polybrene had largest impact at low MOI conditions, but still gave approximately twofold improvement at high MOIs also (e.g., MOI 100). Centrifugation during transduction resulted in significant improvements albeit of comparatively modest magnitude. Finally, two sequential transduction improved efficiency in an approximately additive manner, in the range of 2–2.5-fold above that of a single round of transduction. That the second round of transduction could provide a slightly better rate of transduction than the preceding round (i.e., above what is expected from simple additive outcome of twofold) is intriguing and the mechanism underlying this phenomena require additional investigation. Potentially, cells could be become primed for transduction following initial exposure, transduced cells could act non cell autonomously to improve transduction of un-transduced cells, or cultures could become skewed toward more viral tolerant cells, although we did not see any changes in cell viability or growth in our assays to support the latter. Transfection rates of 20–40% are routinely reported for many cell lines, and hence for many uses, these transduction procedures would suffice. However, the integration and stable expression of the transgene provides means to further purify for transduced cells using selectable markers. Using negative drug selection (blasticidin), we achieved 60% transduction rates two weeks following selection. Using positive FACS based selection, we achieved > 85% transduction rates 2 weeks following selection. The discrepancy between the effectiveness of blasticidin and EGFP based selection (which in our experiments were conducted on expression of an EGFP-blasticidin fusion protein) could be due to the sensitivities of each approach, i.e., that low levels of blasticidin expression (below level of EGFP detection) confers resistance. Other drug-selectable cassettes (e.g., puromycin, neomycin) offer alternative approaches. In any case, drug and fluorescent-based selection methods enriched for transduced cells, but in our hands, the FACS based method provides more utility, as it offers a simple way to isolate live cells that express the highest levels of the transgene.
The ability to efficiently deliver and stably integrate transgenes into LCLs is of great utility. A recent example comes from the EBV field, where the generation of a single stable LCL expressing the expressing the Cas9 nuclease [32] has facilitated CRISPR-Cas9-mediated interrogation of viral factors, host factors, and EBV enhancers including targeted and genome-wide screens [21, 22, 32]. Gene manipulation can in LCLs can open an arsenal of existing and emerging approaches to help to identify and study genetic variation also. The characterisation of new disease-relevant DNA variation in neurological disorders provides an example. Many novel genes and candidate neurological disorder genes may not be expressed in LCLs, however, assessing the effect of a patient’s genetic variation in these cells can still be achieved by, e.g., forced in situ activation of their expression. Lentiviral delivery of the inactive CRISPR-Cas9-VP64 transactivation domain fusion transgene has been developed to activate gene transcription from otherwise silent loci [46], thus allowing the assessment of DNA variation impact (within genomic context) on transcription (e.g., splice-site mutation), on RNA stability (e.g., non-sense mutations), or on protein stability or localisation (e.g., missense mutations). Such tools offer innovative ways to functionally test DNA variants of unknown significance quickly and without great cost. Genetic manipulation of LCLs also offers ways to study disease mechanisms and identify potential disease modifiers. For example, one can test if a potential genetic modifier has an effect by simply overexpressing or silencing the modifier gene and assaying for individual LCL-specific phenotype (e.g., dsyregulated transcription). High throughput formats including genome-wide screens of lentiviral delivered siRNA, or CRISPR guide RNA libraries can be employed to identify modifiers en masse [22, 47, 48]. The functional validation of expression quantitative trait loci, or other genome-wide associations involving the transcriptome, epigenome, proteome, pharmacological sensitivity and beyond, all of which have utilised LCLs, could be tested using CRISPR-based editing [21, 22, 49]. The methods we have described here are translatable across any lentiviral technology that has and will be developed, with both small and large-scale applications, and enables the exciting prospects of combining the vast numbers of genetically diverse LCLs available with genetic manipulation to aid functional interrogation of genotype–phenotype relationships.
References
Neitzel H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum Genet. 1986;73:320–6.
Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–68.
Hussain T, Mulherkar R. Lymphoblastoid cell lines: a continuous in vitro source of cells to study carcinogen sensitivity and DNA repair. Int J Mol Cell Med. 2012;1:75–87.
Mohyuddin A, Ayub Q, Siddiqi S, Carvalho-Silva DR, Mazhar K, Rehman S, et al. Genetic instability in EBV-transformed lymphoblastoid cell lines. Biochim Biophys Acta. 2004;1670:81–83.
Sie L, Loong S, Tan EK. Utility of lymphoblastoid cell lines. J Neurosci Res. 2009;87:1953–9.
International HapMap C, Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–8.
Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74.
Wheeler HE, Dolan ME. Lymphoblastoid cell lines in pharmacogenomic discovery and clinical translation. Pharmacogenomics. 2012;13:55–70.
Caron M, Imam-Sghiouar N, Poirier F, Le Caer JP, Labas V, Joubert-Caron R. Proteomic map and database of lymphoblastoid proteins. J Chromatogr B Anal Technol Biomed Life Sci. 2002;771:197–209.
Dirksen EH, Cloos J, Braakhuis BJ, Brakenhoff RH, Heck AJ, Slijper M. Human lymphoblastoid proteome analysis reveals a role for the inhibitor of acetyltransferases complex in DNA double-strand break response. Cancer Res. 2006;66:1473–80.
Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–10.
Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–26.
Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–11.
Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–5.
Lim ET, Wurtz P, Havulinna AS, Palta P, Tukiainen T, Rehnstrom K, et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494.
Gamazon ER, Duan S, Zhang W, Huang RS, Kistner EO, Dolan ME, et al. PACdb: a database for cell-based pharmacogenomics. Pharm Genom. 2010;20:269–73.
Min JL, Taylor JM, Richards JB, Watts T, Pettersson FH, Broxholme J, et al. The use of genome-wide eQTL associations in lymphoblastoid cell lines to identify novel genetic pathways involved in complex traits. PLoS ONE. 2011;6:e22070.
Welsh M, Mangravite L, Medina MW, Tantisira K, Zhang W, Huang RS, et al. Pharmacogenomic discovery using cell-based models. Pharmacol Rev. 2009;61:413–29.
Izawa K, Martin E, Soudais C, Bruneau J, Boutboul D, Rodriguez R, et al. Inherited CD70 deficiency in humans reveals a critical role for the CD70-CD27 pathway in immunity to Epstein-Barr virus infection. J Exp Med. 2017;214:73–89.
Jiang S, Zhou H, Liang J, Gerdt C, Wang C, Ke L, et al. The Epstein-Barr virus regulome in lymphoblastoid cells. Cell Host Microbe. 2017;22:561–73. e564
Ma Y, Walsh MJ, Bernhardt K, Ashbaugh CW, Trudeau SJ, Ashbaugh IY, et al. CRISPR/Cas9 screens reveal epstein-Barr virus-transformed B cell host dependency factors. Cell Host Microbe. 2017;21:580–91. e587
Chandra S, Levran O, Jurickova I, Maas C, Kapur R, Schindler D, et al. A rapid method for retrovirus-mediated identification of complementation groups in Fanconi anemia patients. Mol Ther. 2005;12:976–84.
Muller LU, Milsom MD, Kim MO, Schambach A, Schuesler T, Williams DA. Rapid lentiviral transduction preserves the engraftment potential of Fanca-/- hematopoietic stem cells. Mol Ther. 2008;16:1154–60.
Portal D, Zhao B, Calderwood MA, Sommermann T, Johannsen E, Kieff E. EBV nuclear antigen EBNALP dismisses transcription repressors NCoR and RBPJ from enhancers and EBNA2 increases NCoR-deficient RBPJ DNA binding. Proc Natl Acad Sci USA. 2011;108:7808–13.
Sommermann TG, O’Neill K, Plas DR, Cahir-McFarland E. IKKbeta and NF-kappaB transcription govern lymphoma cell survival through AKT-induced plasma membrane trafficking of GLUT1. Cancer Res. 2011;71:7291–300.
Wang X, Berger C, Wong CW, Forman SJ, Riddell SR, Jensen MC. Engraftment of human central memory-derived effector CD8 +T cells in immunodeficient mice. Blood. 2011;117:1888–98.
Arvey A, Tempera I, Tsai K, Chen HS, Tikhmyanova N, Klichinsky M, et al. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012;12:233–45.
White RE, Ramer PC, Naresh KN, Meixlsperger S, Pinaud L, Rooney C, et al. EBNA3B-deficient EBV promotes B cell lymphomagenesis in humanized mice and is found in human tumors. J Clin Invest. 2012;122:1487–502.
Banzhaf-Strathmann J, Claus R, Mucke O, Rentzsch K, van der Zee J, Engelborghs S, et al. Promoter DNA methylation regulates progranulin expression and is altered in FTLD. Acta Neuropathol Commun. 2013;1:16.
Zhao B, Barrera LA, Ersing I, Willox B, Schmidt SC, Greenfeld H, et al. The NF-kappaB genomic landscape in lymphoblastoid B cells. Cell Rep. 2014;8:1595–606.
Greenfeld H, Takasaki K, Walsh MJ, Ersing I, Bernhardt K, Ma Y, et al. TRAF1 coordinates polyubiquitin signaling to enhance Epstein-Barr virus LMP1-mediated growth and survival pathway activation. PLoS Pathog. 2015;11:e1004890.
Ohashi M, Holthaus AM, Calderwood MA, Lai CY, Krastins B, Sarracino D, et al. The EBNA3 family of Epstein-Barr virus nuclear proteins associates with the USP46/USP12 deubiquitination complexes to regulate lymphoblastoid cell line growth. PLoS Pathog. 2015;11:e1004822.
Nguyen LS, Jolly L, Shoubridge C, Chan WK, Huang L, Laumonnier F, et al. Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol Psychiatry. 2012;17:1103–15.
Jolly LA, Homan CC, Jacob R, Barry S, Gecz J. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet. 2013;22:4673–87.
Weber K, Bartsch U, Stocking C, Fehse B. A multicolor panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis. Mol Ther. 2008;16:698–706.
Weber K, Mock U, Petrowitz B, Bartsch U, Fehse B. Lentiviral gene ontology (LeGO) vectors equipped with novel drug-selectable fluorescent proteins: new building blocks for cell marking and multi-gene analysis. Gene Ther. 2010;17:511–20.
Koh CM. Preparation of cells for microscopy using cytospin. Methods Enzymol. 2013;533:235–40.
Kumar R, Corbett MA, van Bon BW, Woenig JA, Weir L, Douglas E, et al. THOC2 mutations implicate mRNA-export pathway in X-linked intellectual disability. Am J Hum Genet. 2015;97:302–10.
Corbett MA, Schwake M, Bahlo M, Dibbens LM, Lin M, Gandolfo LC, et al. A mutation in the Golgi Qb-SNARE gene GOSR2 causes progressive myoclonus epilepsy with early ataxia. Am J Hum Genet. 2011;88:657–63.
Maruo S, Wu Y, Ishikawa S, Kanda T, Iwakiri D, Takada K. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc Natl Acad Sci USA. 2006;103:19500–5.
Weber K, Thomaschewski M, Benten D, Fehse B. RGB marking with lentiviral vectors for multicolor clonal cell tracking. Nat Protoc. 2012;7:839–49.
Weber K, Thomaschewski M, Warlich M, Volz T, Cornils K, Niebuhr B, et al. RGB marking facilitates multicolor clonal cell tracking. Nat Med. 2011;17:504–9.
Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10.
Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60.
Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147.
Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–73.
Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554–66.
Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–55.
Acknowledgements
L.A.J. is supported by Australian Research Council DE160100620. J.G. is supported by National Health and Medical Research Council (NHMRC) of Australia grants 1041920 and 1091593. This work was supported by NHMRC GNT1063808 to J.G. and L.A.J.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
This Research was prospectively reviewed and approved by the Women’s and Children’s Hospital Human Research Ethics Committee, South Australia, Australia, 5006. Informed consent was obtained from all individual participants included in the study. The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Jolly, L.A., Sun, Y., Carroll, R. et al. Robust imaging and gene delivery to study human lymphoblastoid cell lines. J Hum Genet 63, 945–955 (2018). https://doi.org/10.1038/s10038-018-0483-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-018-0483-2