Abstract
Mitochondrial aminoacyl-tRNA synthetases (mt-aaRSs) are a family of enzymes that play critical roles in protein biosynthesis. Mutations in mt-aaRSs are associated with various diseases. As a member of the mt-aaRS family, PARS2 encoding prolyl-tRNA synthetase 2 was recently shown to be associated with Alpers syndrome and certain infantile-onset neurodegenerative disorders in four patients. Here, we present two patients in a pedigree with early developmental delay, epileptic spasms, delayed myelination combined with cerebellar white matter abnormalities, and progressive cortical atrophy. Whole-exome sequencing revealed pathogenic compound heterozygous variants [c.283 G > A (p.95 V > I)] and [c.604 G > C (p.202 R > G)] in PARS2. Nearly all patients had epileptic spasms with early response to treatment, early developmental delay and/or regression followed by generalized hypotonia, postnatal microcephaly, elevated lactate levels, and progressive cerebral atrophy. Our study provides further evidence for validating the role of PARS2 in the pathology of related infantile-onset encephalopathy, contributing to the phenotypic features of this condition, and providing clinical and molecular insight for the diagnosis of this disease entity.
Similar content being viewed by others
Introduction
The human nucleus-encoded mitochondrial aminoacyl-tRNA synthetases (mt-aaRSs) constitute a family of 19 enzymes that coordinate nucleic acids and proteins for mitochondrial translation, potentially affecting ATP production in an indirect manner [1]. They catalyze the attachment of an amino acid to its cognate transfer RNA (tRNA) molecule, thus allowing each tRNA to accurately translate the genetic code of DNA into the amino acid code of proteins [2,3,4]. The mt-aaRSs, regarded as “house-keeping” enzymes, play essential and fundamental roles in precisely deciphering the genetic code by charging mitochondrial tRNAs with their cognate amino acids for protein synthesis. Although the basic functions of the mt-aaRSs were established several decades ago, these proteins have recently been shown to be correlated with various genetic diseases including Charcot-Marie-Tooth disease, Perrault syndrome, intellectual disability, leukodystrophies, myopathy, and hearing loss [5,6,7,8,9,10]. Mutations in mt-aaRSs predominantly affect the central nervous system, but other organs may also be involved. As a type of mt-aaRS, prolyl-tRNA synthetase 2(mt-ProRS) encoded by PARS2 is a putative member of the class II family of aminoacyl-tRNA synthetases and catalyzes the ligation of proline to tRNA molecules. Recently, Mizuguchi and Sofou reported that biallelic mutations in PARS2 can cause infantile-onset neurodegenerative disorder and Alpers syndrome [11,12,13]. However, the role of PARS2 in this pathology was only observed in four patients, indicating that additional studies of PARS2 are needed to validate the pathogenicity in relation to the clinical spectrum and determine the potential phenotype-genotype correlations.
Here, we describe the clinical and molecular characterization of a pedigree affected by early developmental delay, epileptic spasms, delayed myelination, with progressive cerebellar white matter abnormalities, and cortical atrophy. By whole-exome sequencing, we identified two pathogenic variants of PARS2 in the family. To delineate the phenotype and describe the cardinal features, we further compared the main clinical and genetic features in patients reported to bear PARS2 mutations. Our findings support the role of PARS2 in the pathology of related infantile-onset encephalopathy, contribute to the phenotypic features of this condition, and provide clinical and molecular insight for the diagnosis of this disease entity.
Materials and methods
Standard protocol approval, registration, and patient consent
All clinical information and materials used in this study were obtained with written informed consent and approved by the Medical Ethics Committee of Xiangya Hospital, Central South University, Changsha, China (IRB [14] NO. 201503487). Written informed consent was obtained from the guardians of the patients. Genomic DNA was prepared from peripheral blood according to standard procedures as previously described [15, 16].
Whole-exome sequencing, variant calling, annotation, and prioritization
Chromosome G-banding staining was performed using cells derived from the two patients. No abnormalities were observed by karyotype analysis. Whole-exome sequencing was performed on the two patients as well as their parents. DNA was isolated using the SureSelect XT library Prep Kit (Agilent Technologies, Santa Clara, CA, USA) and then sequenced on the Illumina HiSeq X Ten platform (Illumina, San Diego, CA, USA) with 150 base pair paired-end reads. BAM and VCF files were attained from the exome data process. Alignment to the human genome assembly hg19 (GRCh37) was carried out, followed by recalibration and variant calling. Single-nucleotide variants and indel calls were considered when these positions had a coverage depth of at least 10 reads. We then used the Clinical Sequence Analysis system from WuxiNextCODE to analyze the variants under autosomal recessive inherited and X-linked inherited models. De novo variations were also considered [17,18,19]. The candidate variants were further confirmed by Sanger sequencing in all available family members.
Results
Clinical characteristics of the family
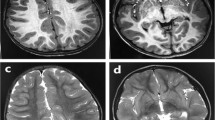
(Summarized in Table 1) The proband, a 3-year-old female, was the first child of healthy, nonconsanguineous parents of Chinese descent (Fig. 1a, II:1). This girl was born at 41 weeks of gestation after an uncomplicated pregnancy and delivery. At birth, her weight, length, and head circumference were within the normal range. However, she showed early signs of developmental alert in the early infantile period. At nearly 1 month of age, she had difficulty feeding from a regular nipple and showed little response to loud sounds and bright lights. At 3 months of age, she could neither focus on moving objects nor lift her head briefly when lying on her stomach. She was able to control her head movement until 6 months of age and sit on her own at nearly 3 years. Since the age of 9 months, she experienced typical epileptic spasms with hypsarrhythmia on electroencephalogram (EEG), indicating a diagnosis of West syndrome (Fig. 1c). The seizures occurred in clusters with a frequency of up to 30 times per day. The patient received antiepileptic treatment with topiramate and sodium valproate. Both the epileptic spasms and hypsarrhythmia on EEG were significantly improved after 2 weeks. One month later, the EEG showed no hypsarrhythmia but multifocal spikes and sharp waves with a symmetric background pattern (Fig. 1c). However, her intellectual disability was profound, and her neurologic examination was notable for generalized hypotonia, dystonia, and progressive microcephaly. Lactate levels in her blood were elevated to 21.7 mg/dL (normal range: 5.0–15.0 mg/dL). Magnetic resonance imaging (MRI) of her brain, performed at 10 months of age, revealed moderate T2 hyperintensities in the subcortical white matter along with decreased frontal lobe volume, and relatively greater T2 signal intensity associated with T1 hypointensity in the bilateral cerebellar white matter around the dentate nuclei (Fig. 1d). At 16 months of age, brain MRI demonstrated blurred gray-white matter interface on the T1-weighted images, suggesting delayed subcortical white matter myelination, along with generalized cortical atrophy (Fig. 1e). Additionally, further progression of the cerebellar white matter abnormalities was observed (Fig. 1f). No signs of cardiac, hepatic, or renal dysfunction were observed. Hearing and visual impairments were not apparent.
Identification of compound heterozygous variants in PARS2 in the studied family. a Pedigree structure of the studied family. Whole exome sequencing was performed for I:1, I:2, II:1, and II:2. The compound heterozygous variants cosegregated with the disease. The genotype is indicated under each family member (–, wild-type allele; v1/v2, variation allele). b Sanger sequencing confirmed the compound heterozygous missense variants [c.283 G > A (p.95 V > I) and c.604 G > C (p.202 R > G)] in affected individuals. c In patient II:1, an EEG at 9 months of age (above) showed typical hypsarrhythmia; an EEG at 10 months of age (below) showed multifocal spikes, sharp waves with a symmetric background pattern without hypsarrhythmia. d In patient II:1, the axial T2-weighted image at 10 months of age showed moderate T2 hyperintensities in the subcortical white matter along with decreased frontal lobe volume (black arrow, d, left), and relatively greater T2 signal intensity associated with T1 hypointensity in the bilateral cerebellar white matter around the dentate nuclei (white arrow, d, middle and right). e In patient II:1, the axial T2-weighted image at 16 months of age displayed mild T2 hyperintensities in the subcortical white matter (e, left), the T1-weighted images show isointensity of the subcortical white matter relative to gray matter, resulting in a blurred gray-white matter interface (e, right). Progressive enlargement of cerebrospinal fluid spaces (white arrowhead, e, right) and subarachnoid space was observed in the bilateral frontal and temporal lobes (black arrow, e, left). f In patient II:1, the axial T2-weighted image at 16 months of age displayed prominent hyperintensities on T2WI and hypointensities on T1WI in expanded lesions of the deep cerebellar white matter (white arrow, f, left and right). g In patient II:2, an EEG at 3 months of age (above) demonstrated typical hypsarrhythmia; an EEG at 4 months of age (below) showed multifocal spikes, polyspike, and sharp waves with an asymmetric background pattern without hypsarrhythmia. h In patient II:2, the axial T2-weighted image at 3 weeks of age showed sight higher T2 intensity in the bilateral cerebellar white matter around the dentate nuclei (white arrow, h)
Individual II, a female, was the second child in this family (Fig. 1a, II:2). She was born after a normal pregnancy and delivered at 40 weeks of gestation. As observed in her older sister (II:1), her weight, length, and head circumference were normal at birth. However, she showed no obvious response to a sound stimulus and less attention to the mother’s face accompanied by poor feeding at a15 days of age. Because her older sister showed abnormal development as a risk factor, an MRI scan of her brain was performed at 3 weeks of age, revealing relatively higher T2 intensity in the bilateral cerebellar white matter around the dentate nuclei (Fig. 1h). Until the age of 3 months, individual II showed little ability to control her head movements. Instead, at this time, she began to develop epileptic spasms with hypsarrhythmia evident by EEG (Fig. 1g), which occurred significantly earlier than in her older sister. The seizures occurred in clusters with a frequency of 20–30 times per day. She was also diagnosed with infantile spasms and treated with adrenocorticotropic hormone, which, unfortunately, showed a less effective treatment response. She still had 5–6 spasms per day after treatment, but hypsarrhythmia also existed for only a very short period (Fig. 1g). Her blood lactate levels increased to 20.3 mg/dL. Hepatic and renal dysfunction were not observed. Visual and auditory evoked potentials were both normal. She died of refractory pneumonia at 4 months of age without achieving further developmental milestones.
Genetic analysis
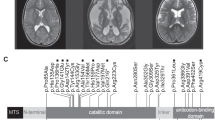
Chromosomal analysis of the two female patients revealed normal female 46, XX karyotypes. Whole-exome sequencing data showed that more than 97% of the target regions had a coverage of at least 10 reads for all four individuals. Through sequencing data sorting, variant filtering, and prioritization, we obtained one candidate gene, PARS2, with compound heterozygous variations. No candidate variants with homozygous, de novo, or X-linked inherited models were found. Sanger sequencing was conducted to confirm cosegregation of the compound heterozygous missense variants (NM_152268, [c.283 G > A (p.95 V > I)]; [c.604 G > C (p.202 R > G)]) in PARS2 with the disorder in this family (Fig. 1b). PARS2 encodes prolyl-tRNA synthetase 2 (mt-ProRS), a mt-aaRSs catalyzing the ligation of proline to tRNA molecules. The paternally inherited variant c.283 G > A (p.95 V > I), previously reported by Mizuguchi et al. in a family with infantile-onset developmental delay/regression and epilepsy, has an allele frequency of 0.0132% in the total of approximately 60,000 individuals in the Exome Aggregation Consortium (ExAC) database and 0.161% in the East Asian population, with no homozygotes identified. This mutation was predicted to be disease-causing by MutationTaster. The other variant, c.604 G > C (p.202 R > G), newly identified in our study, was inherited from the mother, and predicted as “damaging” by SIFT, PolyPhen, and MutationTaster (Table 2). It shows an extremely rare frequency of 1.648E-5 in ExAC and is absent from the dbSNP and 1000 genome databases, which supports the expected very low prevalence of the disease. The R202G mutant changes the evolutionarily conserved arginine to glycine in the class II catalytic motif 2 of the mt-ProRS catalytic domain, which is close to the reported variant c.607 G > A (p.203E > K) (Fig. 2a, b). According to the standards and guidelines for the interpretation of sequence variants from ACMG [17, 20, 21], the compound heterozygous variants that we identified are categorized as “pathogenic variant” belonging to 1 pathogenic strong evidence (PS1), 2 pathogenic moderate evidences (PM1, 3), and ≥ 2 pathogenic supporting evidences (PP1–4), indicating that the variants have high pathogenicity.
Disease-associated mutations of mitochondrial prolyl-tRNA synthetase 2 (mt-ProRS). a The mt-ProRS mutations are shown as a schematic representation; the known functional domains are color-coded. Allelic compositions, as identified in patients, are linked through black lines for all reported mutations. The V95I and R202G mutations identified in this study are indicated in red characters. V95I is in the dimerization domain and R202G is in the class II catalytic motif 2 domain. b Evolutionary conservation of p.Val95 and p.Arg202 amino acid residues in the mt-ProRS reveals that these residues are highly conserved across several species
Discussion
We identified pathogenic compound heterozygous variations in PARS2 in the two patients with early developmental delay, epileptic spasms, delayed myelination combined with cerebellar white matter lesion, and progressive cortical atrophy by exome sequencing of parent-child trios. Compound heterozygous mutations were first reported in PARS2 in a patient presenting with infantile mitochondrial Alpers encephalopathy in 2015 [12]. In 2016 and 2017, two studies describing the pathogenic role of PARS2 biallelic variants in infantile-onset developmental delay/regression and epilepsy were published [11, 13]. Here, we describe the similarities and differences between the affected sisters in China and cases presented in previous studies (Table 1) in hopes of providing insight into the main clinical features and phenotype-genotype relationships of the disease. Unlike some other mt-aaRS enzymes, mutations in mt-ProRS lead exclusively to epileptic encephalopathy. The phenotypic spectrum of PARS2-related infantile-onset encephalopathy shows wide variation and heterogeneity, as reflected by the range of clinical characteristics summarized in Table 1. However, a core set of clinical features shared by our patients has emerged, including epilepsy, characterized by epileptic spasms with short duration, early developmental delay and/or developmental regression followed by generalized hypotonia, postnatal microcephaly, elevated lactate levels, and progressive cerebral atrophy. Other phenotypes are also observed in these patients, such as cardiac anomaly, cortical visual impairment, renal dysfunction, hyperreflexia, and cerebral white matter hypomyelination. Compared to individuals with NARS2 (asparaginyl-tRNA synthetase 2) mutations, patients with PARS2 mutations may have no obvious sensorineural hearing impairments. Moreover, unlike the myoclonic seizures associated with NARS2 mutations, early onset epileptic spasms with or without hypsarrhythmia appear to be a common feature of PARS2-related infantile-onset encephalopathy. Interestingly, hypsarrhythmia on EEG occurs over a short period and the epileptic spasms tend to be responsive to anti-epileptic drugs in nearly all patients. Although the epileptic spasms are easily controlled, the intellectual disability and psychomotor regression were still profound. In some patients, focal impaired awareness seizures were observed at later ages, also indicating progressive neurodegeneration. Activities of complex I and IV in the mitochondria may be decreased or within the normal range. However, hepatic dysfunction has not been reported, which differs from Alpers syndrome caused by mutations in POLG1. Of these six patients, three have died at infancy or during childhood, mainly because of heart failure and/or respiratory dysfunction. Some patients can survive to school age.
Compared to the sisters with similar mutations reported by Mizuguchi, two of our patients showed earlier developmental delay with feeding difficulties. Moreover, the younger sister suffered from respiratory dysfunction that led to early death. Another specific clinical manifestation of our patients was the delayed myelination in subcortical white matter combined with progressive cerebellar white matter abnormalities, which is significantly different from MRI findings in previously reports [11, 12]. Delayed myelination maybe a common feature observed in children with delayed development [22]. It is also possible that there are superimposed lesions on a background of diffuse delayed myelination/deficient myelination [23, 24], indicating instability of myelin formation and maintenance. In early onset neuronal degenerative disorders, deficient myelination may also be more inhomogeneous and patchy. The prominent changes of cerebellar white matter in our patients may represent myelin damage caused by the loss of oxygen and/or metabolic problems in mitochondrial disorders. The cerebellum is sensitive and often the target of mitochondrial dysfunction. In patients with Alpers syndrome, the cerebellum, basal ganglia, and thalamus were involved to variable extents, and at least 21% (4/19) patients were with cerebellar lesions in neuroimaging. Neuropathological changes in the cerebellum were also observed in most patients with Alpers syndrome, and the types of alterations were inconsistent. Focal loss of Purkinje cells with gliosis and spongiosis in dentate nucleus and white matter were found [25]. Additionally, the patient with PARS2 mutations reported by Ewa Pronicka showed Leigh-like syndrome with basal ganglia involvement [13], supporting that sensitive regions, such as the basal ganglia, thalamus, and cerebellum, are damaged by oxygen and metabolic problems in this disorder.
The exact pathogenesis of cerebellar white matter changes require further exploration and discussion. It has been suggested that cerebellar white matter lesions lead to hypoactivity in supratentorial areas of the brain via disconnection of cortical areas or disruption of the cerebello-cerebral circuitry, resulting in cognitive and language dysfunction [26, 27], which may contribute to the pathologic basis of our patients. Because PARS2-related syndromes exhibit variable phenotypic features and clinical heterogeneity, it is likely that our newly described MR finding is inconsistent with previous observations. This is important because cerebellar white matter abnormalities appear early in the course of disease and may help to early diagnosis of the disorders.
The variant c.283 G > A (p.95 V > I) identified in this study was located on a highly conserved dimerization domain in PARS2 but predicted to be benign by partial prediction tools. The frequency of this variant was 0.001619 in the East Asian population but rare or absent in other populations in the ExAC, indicating that it acts as a hypomorphic allele that contributes to a severe neurological phenotype only when in trans with a deleterious mutation in PARS2. This relatively high frequency may be a result of either a founder effect or selective advantage for heterozygotes. While this variant in PARS2, with a deleterious variant in trans, was identified in four patients from two unrelated families with similar encephalopathy phenotypes, the data suggests that even mildly impaired function from recessive mutations can cause mitochondrial disease [11]. Such pathogenic variants with high ancestry-specific minor allele frequency are not rare in disorders inherited as autosomal recessive traits. For instance, the carrier frequency for the 35delG deafness mutation in GJB2 was 1 in 51 in the overall European population [28].
To date, including those in our study, seven pathogenic variations in PARS2 have been identified in four separate families [11,12,13]. All variations were compound heterozygotes (Fig. 2a) and accounted for ~4.5% (7/154) of the total mutations in nuclear-DNA encoded mt-aaRSs [10]. While these variants are found throughout the constitutive functional domains of mt-ProRS, three were positioned in the core catalytic domain, strongly suggesting that the affected function of this core domain is involved in disease generation [11, 12]. The only frameshift mutation was in the anticodon binding domain, suggesting a direct impact on aminoacylation function [10]. More importantly, the clinical manifestations of patients with compound heterozygous missense variants were less severe than those of patients carrying a missense variant combined with a frameshift mutation. This indicates that the pathogenicity of mutated mt-ProRS exclusively relies on the retained activity of the enzyme. Because disease-related mutations in mt-aaRSs do not lead to multi systemic injuries and the central nervous system is the most frequently affected system, numerous specific mechanisms may affect, exploit, or modulate the mitochondrial translation apparatus in neuronal cells.
Taken together, we identified pathogenic variants of PARS2 in two patients with early developmental delay, infantile spasm, delayed myelination combined with cerebellar white matter abnormalities, and progressive cortical atrophy. Although a broad range of clinical characteristics is associated with PARS2 mutations, a core set of phenotypic features of PARS2-related infantile-onset encephalopathy can be summarized. Clinicians should consider including this condition in the differential diagnoses suggested by features such as epileptic spasms with early response to treatment, early developmental delay and/or regression followed by generalized hypotonia, postnatal microcephaly, elevated lactate levels, and progressive cerebral atrophy.
References
Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet. 2011;45:299–329.
Havrylenko S, Mirande M. Aminoacyl-tRNA synthetase complexes in evolution. Int J Mol Sci. 2015;16:6571–94.
Francklyn C, Musier-Forsyth K, Martinis SA. Aminoacyl-tRNA synthetases in biology and disease: new evidence for structural and functional diversity in an ancient family of enzymes. RNA. 1997;3:954–60.
Woese CR, Olsen GJ, Ibba M, Soll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64:202–36.
Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72:1293–9.
Mazurova S, Magner M, Kucerova-Vidrova V, Vondrackova A, Stranecky V, Pristoupilova A, et al. Thymidine kinase 2 and alanyl-tRNA synthetase 2 deficiencies cause lethal mitochondrial cardiomyopathy: case reports and review of the literature. Cardiol Young-. 2016;27:936–44.
Simon M, Richard EM, Wang X, Shahzad M, Huang VH, Qaiser TA, et al. Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh syndrome. PLoS Genet. 2015;11:e1005097.
Antonellis A, Green ED. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu Rev Genom Hum Genet. 2008;9:87–107.
Konovalova S, Tyynismaa H. Mitochondrial aminoacyl-tRNA synthetases in human disease. Mol Genet Metab. 2013;108:206–11.
Sissler M, Gonzalez-Serrano LE, Westhof E. Recent advances in mitochondrial aminoacyl-tRNA synthetases and disease. Trends Mol Med. 2017;23:693–708.
Mizuguchi T, Nakashima M, Kato M, Yamada K, Okanishi T, Ekhilevitch N, et al. PARS2 and NARS2 mutations in infantile-onset neurodegenerative disorder. J Hum Genet. 2017;62:525–9.
Sofou K, Kollberg G, Holmstrom M, Davila M, Darin N, Gustafsson CM, et al. Whole exome sequencing reveals mutations in NARS2 and PARS2, encoding the mitochondrial asparaginyl-tRNA synthetase and prolyl-tRNA synthetase, in patients with Alpers syndrome. Mol Genet Genom Med. 2015;3:59–68.
Pronicka E, Piekutowska-Abramczuk D, Ciara E, Trubicka J, Rokicki D, Karkucinska-Wieckowska A, et al. New perspective in diagnostics of mitochondrial disorders: two years’ experience with whole-exome sequencing at a national paediatric centre. J Transl Med. 2016;14:174.
Zweig RM, Hedreen JC, Jankel WR, Casanova MF, Whitehouse PJ, Price DL, et al. Pathology in brainstem regions of individuals with primary dystonia. Neurology. 1988;38:702–6.
Wang JL, Cao L, Li XH, Hu ZM, Li JD, Zhang JG, et al. Identification of PRRT2 as the causative gene of paroxysmal kinesigenic dyskinesias. Brain. 2011;134:3493–501.
Wang JL, Yang X, Xia K, Hu ZM, Weng L, Jin X, et al. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain. 2010;133:3510–8.
Mao X, Li K, Tang B, Luo Y, Ding D, Zhao Y, et al. Novel mutations in ADSL for Adenylosuccinate Lyase Deficiency identified by the combination of Trio-WES and constantly updated guidelines. Sci Rep. 2017;7:1625.
Ding D, Chen Z, Li K, Long Z, Ye W, Tang Z, et al. Identification of a de novo DYNC1H1 mutation via WES according to published guidelines. Sci Rep. 2016;6:20423.
Chen Z, Ye W, Long Z, Ding D, Peng H, Hou X, et al. Targeted next-generation sequencing revealed novel mutations in chinese ataxia telangiectasia patients: a precision medicine perspective. PLoS One. 2015;10:e0139738.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Li Q, Wang K. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100:267–80.
van der Knaap MS, Valk J, Bakker CJ, Schooneveld M, Faber JA, Willemse J, et al. Myelination as an expression of the functional maturity of the brain. Dev Med Child Neurol. 1991;33:849–57.
Schiffmann R, van der Knaap MS. Invited article: an MRI-based approach to the diagnosis of white matter disorders. Neurology. 2009;72:750–9.
Steenweg ME, Vanderver A, Blaser S, Bizzi A, de Koning TJ, Mancini GM, et al. Magnetic resonance imaging pattern recognition in hypomyelinating disorders. Brain. 2010;133:2971–82.
Sofou K, Moslemi AR, Kollberg G, Bjarnadottir I, Oldfors A, Nennesmo I, et al. Phenotypic and genotypic variability in Alpers syndrome. Eur J Paediatr Neurol. 2012;16:379–89.
Stoodley CJ, Schmahmann JD. The cerebellum and language: evidence from patients with cerebellar degeneration. Brain Lang. 2009;110:149–53.
van Baarsen KM, Grotenhuis JA. The anatomical substrate of cerebellar mutism. Med Hypotheses. 2014;82:774–80.
Gasparini P, Rabionet R, Barbujani G, Melchionda S, Petersen M, Brondum-Nielsen K, et al. High carrier frequency of the 35delG deafness mutation in European populations. Genetic Analysis Consortium of GJB2 35delG. Eur J Hum Genet. 2000;8:19–23.
Acknowledgements
We are indebted to all the patients and family members for their generous participation in this work. We also thank all the clinicians (including pediatricians), laboratory scientists, and bioinformaticians for discussion on the interpretation of sequencing variants. Many thanks go in particular to Ph.D. Ananya Ray-Soni at the Massachusetts General Hospital, Harvard Medical School and Ph.D. Arif Muhammad at Center for Medical Genetics, Central South University, for their suggestions, language editing and proofreading. This work was supported by the National Natural Science Foundation of China [grant numbers 81300980, 81130021, and 81601119].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Yin, X., Tang, B., Mao, X. et al. The genotypic and phenotypic spectrum of PARS2-related infantile-onset encephalopathy. J Hum Genet 63, 971–980 (2018). https://doi.org/10.1038/s10038-018-0478-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-018-0478-z
This article is cited by
-
Genome-wide association study for single nucleotide polymorphism associated with mural and cumulus granulosa cells of PCOS (polycystic ovary syndrome) and non-PCOS patients
Future Journal of Pharmaceutical Sciences (2023)
-
Role of Mutations of Mitochondrial Aminoacyl-tRNA Synthetases Genes on Epileptogenesis
Molecular Neurobiology (2023)
-
Four pedigrees with aminoacyl-tRNA synthetase abnormalities
Neurological Sciences (2022)