Abstract
X-linked inheritance is very rare and is estimated to account for only 1–5% of all nonsyndromic hearing loss cases. We found a multiplex family from China segregating with X-linked nonsyndromic hearing loss. After exclusive analysis of 10 common variations of three hearing loss-related genes, GJB2, mtDNA12srRNA and SLC26A4, a novel truncated variant of SMPX, c.87dupA (p.Gly30Argfs*12) (NCBI ClinVar Submission ID: SUB3136126), was identified by whole-exome sequencing. This variant was co-segregated with hearing loss in the entire family and was absent in 576 unrelated ethnically and geographically matched controls. We also detected a single nucleotide variation in two male controls with normal hearing, SMPX c.55A>G (p.Asn19Asp), which has been annotated as a rare variant in the Single Nucleotide Polymorphism (dbSNP) (rs759552778) and Exome Aggregation Consortium (ExAC) databases. This study has enriched the mutation spectrum of the SMPX gene.
Similar content being viewed by others
Introduction
Hearing loss (HL) affects 15–26% of the world’s population and 2–3 of every 1000 newborns [1]. Both genetic and environmental factors can lead to HL, and more than half of congenital HL cases can be traced to a genetic cause. The majority of inherited HL is nonsyndromic (nonsyndromic hearing loss, NSHL) and is related to 109 reported genes, most of which are located in autosomes. X-linked inheritance is very rare and is estimated to account for <1% of all cases of NSHL worldwide [2]. Male patients often experience earlier onset with more severe presentation than females due to X inactivation [3]. Currently, there are six known loci for X-linked HL, and five genes have been identified: DFNX1:PRPS1 (OMIM 304500) [4], DFNX2:POU3F4 (OMIM 304400) [5], DFNX3:unknown gene [6] (OMIM 300030), DFNX4:SMPX (OMIM 300066) [7], DFNX5:AIFM1 (OMIM 300614) [8, 9] and DFNX6:COL4A6 (OMIM 303630) [10].
SMPX (RefSeq NM_014332) is the only gene responsible for X-linked dominant NSHL, and six pathogenic mutations of SMPX have been shown to lead to deafness [11]. Heart and skeletal muscle express SMPX preferentially and abundantly [12], and various cell types of the murine cochlea also express SMPX[13]. The stress of mechanical challenges innate to hearing have been suggested to damage certain cells of the inner ear due to mutated Smpx [13]. The six known pathogenic mutations of the SMPX gene are located in its second and third exons, leading to the production of truncated proteins [7, 11,12,13,14].
Recently, the application of whole-exome sequencing (WES) has advanced dramatically, which has greatly helped in the identification of causative genes [15, 16]. In this study, using WES, we identified a truncated mutation, SMPXc.87dupA, in a multiplex, extended Chinese pedigree that co-segregated with X-linked sensorineural HL, thus enriching the mutation spectrum of SMPX in Chinese people.
Materials and methods
Subjects and clinical assessment
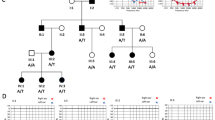
We investigated X-linked dominant inheritance in a four-generation family from Hunan Province, China (Fig. 1), with a total of 59 people, including 12 patients. All participants were given a general physical as well as otological examinations by two experienced otologists independently. An otoscope was used to exclude otitis media and perforation. Pure-tone audiometry (PTA) and acoustic immittance measurements were used to assess the function of the middle ear and tympanic membrane as well as hearing sensitivity. Auditory brainstem response (ABR), multiple frequency auditory steady-state evoked responses (ASSR), distortion product otoacoustic emission (DPOAE) and vestibular testing were performed on specific individuals. To exclude syndromes of other organs, the proband (4-3) and his mother (3-6) each received an ophthalmic examination, routine blood examination, routine urine test, routine stool test, temporal bone high-resolution computed tomography (HRCT) and brain magnetic resonance imaging (MRI). The classification criteria used for HL was as follows: (<25 dB HL) normal, (26–40 dB HL) mild/slight, (41–60 dB HL) moderate, (60–80 dB HL) severe and (>81 dB HL) profound. The controls consisted of 576 unrelated Chinese healthy volunteers with normal hearing (308 males and 268 females; the minimum age of the males was 35, and the minimum age of the females was 42). All patients and controls were ethnically Chinese, with no parents or grandparents of any ethnicity other than Chinese.
Pedigree of X-linked nonsyndromic sensorineural hearing loss family CD-049. Open symbols, normal hearing; black solid symbols, hereditary hearing loss; and grey solid symbols, nonhereditary hearing loss. Squares, males; circles, females; slanting lines, deceased subjects; slanting arrow, the proband; and question mark, phenotype unknown
This study was approved by the Medical Ethics Committee at Central South University Xiangya Hospital. All participants in the study provided the written informed consent.
Exclusive analysis
Mutational analysis of the proband was performed for 10 common variations of three HL-related genes: GJB2, mtDNA 12S rRNA and SLC26A4 (GJB2 c.235delC, c.299-300delAT, c.176dell bp, and c.512insAAGG; SLC26A4 IVS7-2A>G, c.2168A>G, c.1229C>T and c.1174A>T; and 12S rRNA A1555G and C1494T) using fluorescent polymerase chain reaction (PCR).
WES and bioinformatic analysis
DNA was extracted from peripheral blood using standard phenol–chloroform protocols and was then stored at −20 °C. Six micrograms of the proband’s DNA was sent to the Annoroad Gene Technology Co. Ltd. in Beijing, China, for WES. The DNA sample was sheared by sonication, yielding an average DNA size of ~200 bp. The DNA was purified and end-repaired with an adenine base added to each fragment, and the paired-end adaptor was then ligated and amplified to yield each DNA library. The sample and target-specific capture libraries were hybridized and washed, followed by recovery of the captured DNA sample. Exon-enriched DNA was sequenced using an Illumina HiSeq2500 platform following the manufacturer’s instructions. Raw image files were processed using BclToFastq for base calling and raw data generation. Low-quality variations were filtered out using a quality score ≥30 (Q30). The sequencing reads were aligned to the NCBI human reference genome (hg19) using the Burrows–Wheeler Aligner (BWA). Samtools and Pindel were used to analyze single nucleotide polymorphisms (SNPs) and indels in the sequence data. Synonymous changes and SNPs with minor allele frequencies (MAFs) above 5% were removed (http://www.ncbi.nlm.nih.gov/projects/SNP). Nonsynonymous changes were filtered using SIFT software (http://sift.jcvi.org). In the progressive data analysis, first, the mutation data for exons and splice sites were collected. Mutations with frequencies of less than 0.01 in the 1000 Genomes project were then detected in the filtered data. Subsequently, mutations with frequencies of less than 0.01 in the Exome Aggregation Consortium East Asian subpopulation (ExAC_EAS) database were detected in the filtered data. The selected mutations were then compared to sequences in the known deafness gene database to determine known deafness gene mutations. In the fifth step, the SMPX gene mutation was identified by excluding the incompatible genes according to X-linked dominant inheritance.
Sanger sequencing
Sanger sequencing of SMPX (RefSeq NM_014332) was performed for all family members to determine if the gene co-separated with the HL phenotype in the family. In addition, the 576 control individuals were subjected to Sanger sequencing to exclude benign polymorphism. The PCR products were sequenced on an ABI 3730XL DNA analyzer and evaluated using Lasergene software.
Results
Clinical features
The affected subjects from family CD-049 showed X-linked NSHL. The proband was a 22-year-old boy. His onset age was seven, and his hearing ability continued to diminish in later years, developing into severe sensorineural HL in both ears. As shown in Table 1, the onset age of male patients was seven, which is earlier than that of the female patients, whose onset age was at least 30 years old. HL was more serious in males than in females. Males exhibited fast progressive HL until the age of 20, which then developed into severe sensorineural HL. The hearing of females declined slowly and then developed into severe HL at 45–50 years, with their HL appearing to affect high and middle frequencies to all frequencies. There was no effect from acquired factors such as noise exposure, otogenic trauma, ototoxic medication or ear infections. Patients had not had tinnitus, vestibular dysfunction or muscle abnormalities. No ear abnormalities were detected via HRCT or other tests in subjects 3-6 and 4-3.
All patients exhibited sensorineural HL with binaural symmetrical decrease, except for subject 3-4 who exhibited severe HL in the right ear and normal hearing in the left ear. The symptoms of the majority of patients were similar, except for subject 3-10 who exhibited binaural pus at age 10, a symptom that reappeared over the next several decades. Physical examination revealed bilateral tympanic membrane perforation and white epithelioid matter in the middle ear cavity. PTA revealed that both ears had moderately conductive HL. HRCT showed cholesteatoma in both middle ears. The genetic tests revealed that she did not carry SMPX mutations, and none of her offspring were deaf or carried the SMPX mutation. Thus, we diagnosed subject 3-10 with HL due to bilateral chronic otitis media with cholesteatoma, not hereditary hearing loss (HHL), and her data were excluded from the family analysis. Representative audiograms of the family members are shown in Fig. 2.
Pure-tone audiograms of four patients with HL in the family. a, b, and c are audiograms showing sensorineural hearing loss, while that of d shows conductive deafness. The air conduction results are denoted by a blue “x” for the left ear and a red “o” for the right ear. The bone conduction results are denoted as a blue “>” for the left ear and a red “<” for the right ear. Blue arrows slanting downwards and red arrows slanting downwards denote no response from the left and right ear, respectively
Analysis of WES data
A total of 5.2 Gb of raw WES data were obtained. After the raw data were filtered, 61,236 SNPs and 7954 INDELs were identified. As shown in Table 2, a total of 18,565 mutations in exonic and splice site regions were ascertained from the SNP and INDEL data. Then, 1707 mutations were selected based on their mutation frequency of no more than 1% in the 1000 Genomes project database (http://www.internationalgenome.org/home). Subsequently, a second set of 640 mutations were selected based on their mutation frequency of no more than 1% in the ExAC_EAS database. We selected LOXHD1, MYO7A and SMPX mutations after comparing these mutation sets to known nonsyndromic deafness gene data (http://hereditaryhearingloss.org/). In combination with family genetic analysis, we found that only the SMPX gene is located on the X chromosome and conforms to X-linked dominant genetic characteristics. The variant c.87dupA:p.Gly30Argfs*12 of the SMPX gene (https://submit.ncbi.nlm.nih.gov/subs/variation_clinvar/SUB3136126/) was preliminarily believed to be responsible for HL in the family.
Sanger sequencing in the family and controls
Sanger sequencing of SMPX was performed in all family members to determine whether this gene co-separated with the HL phenotype. Sanger sequencing showed that the SMPX variant c.87dupA co-segregated with HL in the family (Fig. 3a–c). We further confirmed the absence of this SMPX variant in 576 unrelated ethnically and geographically matched controls. In two male controls with normal hearing, we detected a single nucleotide variation in SMPX, c.55A>G:(p.Asn19Asp) (Fig. 3d), which has been annotated as a rare variant in the dbSNP (rs759552778) and ExAC databases.
Genetic map of the SMPX gene and two variations found in the study compared to six pathogenic mutations. aSMPX gene in the Xp21.12 chromosomal region (red rectangle). bSMPX gene mutations. The six known mutations are shown in black, and the variations identified in this study are shown in green and red. c Sequencing diagram of SMPX showing the truncation mutation c.87dupA:(p.Gly30Argfs*12) found in this study. The mutated location is indicated by red arrows. d Sequencing diagram of SMPX showing the missense mutation c.55A>G found in this study. The mutated location is indicated by red arrows. e Two mutations corresponded to predicted protein sequences with known pathogenic mutations in this study. The red vertical line indicates amino acids that differ from those of the reference protein, while the red rectangles show the changed amino acids
Discussion
Five X-linked deafness genes have been identified. Male patients with PRPS1 mutations show prelingual or postlingual, bilateral, moderate to profound, progressive or nonprogressive HL, while female patients show unilateral or bilateral HL [4, 17]. The clinical characteristics of POU3F4 mutations in affected males include temporal bone abnormalities, stapes fixation, and in most cases, a mixed type of HL in which the patients typically present with temporal bone abnormalities and stapes fixation [18]. The HL with these mutations is often progressive. Female patients with POU3F4 mutations typically show little or no HL [19,20,21]. In male patients, COL4A6 mutations may cause bilateral severe sensorineural HL and cochlear malformation, whereas female patients show no HL or only mild to moderate sensorineural HL [10]. AIFM1 gene mutations can lead to X-linked recessive auditory neuropathy spectrum disorder [8, 9], the phenotype of which associated with heterozygous males is characterized by childhood-onset auditory neuropathy spectrum disorder and delayed peripheral sensory neuropathy and presents as extremity numbness, unsteadiness and reflexia. Heterozygous females have no phenotype. HL caused by SMPX gene mutations is significantly different from that caused by the above four gene mutations.
The human SMPX gene is located in chrXp22.12 and consists of five exons. The coding sequence is 267 bp and is composed of part of exon 2, all of exon 3 and part of exon 4 (Fig. 3a, b). The human SMPX protein is composed of 88 amino acids [12] and has no known functional domains. To date, six mutations in the SMPX gene associated with X-linked NSHL have been identified. In 2011, Huebner et al. [13] identified two pathogenic mutations using linkage analysis: SMPX c.175G>T:p.Gly59X in a German pedigree and SMPX c.109G>T:p.Glu37X in a Spanish family. Among these two families, the onset of HL in male carriers is 3–7 years; the patients first present moderate HL for high frequencies only, and HL then begins to affect all frequencies with more advanced age. The onset of HL in female patients occurs in the second to third decades, and they present with severe HL after 10–15 years. Schraders et al. [7] also identified two pathogenic SMPX mutations in 2011, SMPX c.214G>T:p.Glu72X and SMPX c.130delG:(p.Glu44Argfs*37), in two Dutch families using next-generation sequencing. In male patients in these families, fast HL occurred in their first two decades, with progression to profound HL in their second decade. In female carriers, HL exhibited large interindividual variation with regard to severity and interaural variation. In 2012, Abdelfatah et al. [14] identified a SMPX pathogenic mutation, SMPX c.99delC:p.Arg34Glufs*47, in a Newfoundland family via a genotype scan of the X chromosome of four affected family members and one unaffected spouse. He found that the phenotype caused by the mutation was similar to that of the above study. In 2017, we identified, for the first time, a novel pathogenic mutation of the SMPX gene, SMPX c.217dupA:(p.Ile73Asnfs*5), in a Chinese family using WES [11]. The two male babies in the family showed prelingual deafness. Interestingly, all six of these known pathogenic mutations result in truncated proteins (Fig. 3e), suggesting that the poor function of the truncated protein leads to HL. In this study, an X-linked dominance pattern of inheritance was detected based on early onset in the absence of male-to-male transmission as well as a more severe phenotype in males than in females (Fig. 4). The symptoms are similar to those of the above families. This novel variant, SMPX c.87dupA (p.Gly30Argfs*12), co-segregated with the HL phenotype in the family and led to the generation of truncated proteins. MutationTaster predicted that part of SMPX is transcribed and translated, leading to the second shortest SMPX truncated protein compared to the normal integrated SMPX protein (Fig. 3e). Truncated mutations may lead to early mRNA metabolism, resulting in insufficient SMPX protein function.
Hearing threshold of male patients and female patients in the pedigree, males’ phenotype is more severe than females’. The threshold is the average hearing threshold of the left and right ears tested in the study begin. The blue full line shows the male patients, the red dotted line shows the female patients, excluding nonhereditary deaf patient 3-10
Within adult mouse striated muscle cells, SMPX is expressed in myofilaments and costameres, which are subsarcolemmal sites of cytoskeletal or membrane adhesion complexes [22]. Studies found that Smpx may participate in the regulation of cytoskeletal dynamics through the Rac1–p38 pathway in muscle cells [23]. SMPX stained intracellular HeLa cells, with enrichment mainly in lamellipodia, and SMPX and vinculin partially overlap in the peripheral adhesion complex. Smpx was found to be expressed in the mouse inner ear at 14.5 days of gestation and in the human inner ear at 8 weeks of gestation. Mouse cochlear immunostaining revealed that SMPX was expressed in various cells, including pillar cells, root cells, interdental cells and Böttcher cells of the limbus spiralis. Low immune SMPX activity was detected in hair cells [13]. Many genes associated with HL encode actin, actin-binding proteins, cytoskeleton proteins or motor proteins of the myosin family. Given the detection of Smpx in mouse hair cells, the association of SMPX with the cytoskeleton and its responsiveness to mechanical force, SMPX might play an important role in the protection of stereocilia, which are permanently exposed to physical forces. Thus, Smpx might have a protective role against mechanical stress in some cell types of the Corti during the process of hearing [13]. Truncated Smpx protein would not protect the cochlea from chronical mechanical stress and therefore can cause HL.
In our previous study, we identified an SMPX variant, c.55A>G, in sporadic HL patients that was predicted to be functionally deleterious and was not found in 295 normal-hearing control patients from China. In this study, we detected this variation in two male controls with normal hearing (2/576). The variant SMPX c.55A>G led to a change in the 19th amino acid from asparagine to aspartic acid, but the predicted length of the protein did not change (Fig. 3e). Asparagine and aspartic acid are similar in general structure. The function of the mutant protein is likely not significantly decreased. The MAF of this variant is 0.0001702 in the gnomAD database (http://gnomad.broadinstitute.org/), and the allele frequency is 0.0023697 (1/422) in Chinese people, proving that it may be a benign polymorphism.
In conclusion, this study identified a novel SMPX mutation, c.87dupA (p. Gly30Argfs*12), using WES. This pathogenic mutation is transcribed and translated into the second shortest Smpx truncated protein known to date. The symptoms of the pathogenic mutation were characterized by an X-linked dominant inheritance pattern, sensorineural HL with early onset, normal morphological cochlea and more severe phenotypes in males. The comprehensive analysis method used herein, which included the comparison of WES results to known deafness genes as well as a family genetic characteristics analysis, will help to identify very rare HHL gene mutations and novel HHL genes. Our results have enlarged the mutation spectrum of the SMPX gene and will benefit genetic counselling based on mutation analysis for affected families.
References
Yan D, Liu XZ. Cochlear molecules and hereditary deafness. Front. Biosci. 2008;13:4972–83.
Petersen MB, Wang Q, Willems PJ. Sex-linked deafness. Clin Genet. 2008;73:14–23.
Dobyns WB. The pattern of inheritance of X-linked traits is not dominant or recessive, just X-linked. Acta Paediatr Suppl. 2006;95:11–5.
Mittal R, Patel K, Mittal J, Chan B, Yan D, Grati M, et al. Association of PRPS1 mutations with disease phenotypes. Dis Markers. 2015;2015:127013.
de Kok YJ, van der Maarel SM, Bitner-Glindzicz M, Huber I, Monaco AP, Malcolm S, et al. Association between X-linked mixed deafness and mutations in the POU domain gene POU3F4. Science. 1995;267:685–8.
Lalwani AK, Brister JR, Fex J, Grundfast KM, Pikus AT, Ploplis B, et al. A new nonsyndromic X-linked sensorineural hearing impairment linked to Xp21.2. Am J Hum Genet. 1994;55:685–94.
Schraders M, Haas SA, Weegerink NJ, Oostrik J, Hu H, Hoefsloot LH, et al. Next-generation sequencing identifies mutations of SMPX, which encodes the small muscle protein, X-linked, as a cause of progressive hearing impairment. Am J Hum Genet. 2011;88:628–34.
Wang QJ, Li QZ, Rao SQ, Lee K, Huang XS, Yang WY, et al. AUNX1, a novel locus responsible for X linked recessive auditory and peripheral neuropathy, maps to Xq23-27.3. J Med Genet. 2006;43:e33–e33.
Zong L, Guan J, Ealy M, Zhang Q, Wang D, Wang H, et al. Mutations in apoptosis-inducing factor cause X-linked recessive auditory neuropathy spectrum disorder. J Med Genet. 2015;52:523–31.
Rost S, Bach E, Neuner C, Nanda I, Dysek S, Bittner RE, et al. Novel form of X-linked nonsyndromic hearing loss with cochlear malformation caused by a mutation in the type IV collagen gene COL4A6. Eur J Human Genet. 2014;22:208–15.
Niu Z, Feng Y, Mei L, Sun J, Wang X, Wang J, et al. A novel frameshift mutation of SMPX causes a rare form of X-linked nonsyndromic hearing loss in a Chinese family. PLoS ONE. 2017;12:e0178384.
Patzak D, Zhuchenko O, Lee CC, Wehnert M. Identification, mapping, and genomic structure of a novel X-chromosomal human gene (SMPX) encoding a small muscular protein. Hum Genet. 1999;105:506–12.
Huebner AK, Gandia M, Frommolt P, Maak A, Wicklein EM, Thiele H, et al. Nonsense mutations in SMPX, encoding a protein responsive to physical force, result in X-chromosomal hearing loss. Am J Hum Genet. 2011;88:621–7.
Abdelfatah N, Merner N, Houston J, Benteau T, Griffin A, Doucette L, et al. A novel deletion in SMPX causes a rare form of X-linked progressive hearing loss in two families due to a founder effect. Hum Mutat. 2013;34:66–9.
Wang H, Wang X, He C, Li H, Qing J, Grati M, et al. Exome sequencing identifies a novel CEACAM16 mutation associated with autosomal dominant nonsyndromic hearing loss DFNA4B in a Chinese family. J Hum Genet. 2015;60:119–26.
Cai XZ, Li Y, Xia L, Peng Y, He CF, Jiang L, et al. Exome sequencing identifies POU4F3 as the causative gene for a large Chinese family with non-syndromic hearing loss. J Hum Genet. 2017;62:317–20.
Liu XZ, Xie D, Yuan HJ, de Brouwer AP, Christodoulou J, Yan D. Hearing loss and PRPS1 mutations: Wide spectrum of phenotypes and potential therapy. Int J Audiol. 2013;52:23–8.
Stanton SG, Griffin A, Stockley TL, Brown C, Young TL, Benteau T, et al. X-linked hearing loss: two gene mutation examples provide generalizable implications for clinical care. Am J Audiol. 2014;23:190–200.
Bademci G, Lasisi A, Yariz KO, Montenegro P, Menendez I, Vinueza R, et al. Novel domain-specific POU3F4 mutations are associated with X-linked deafness: examples from different populations. BMC Med Genet. 2015;16:9.
Du W, Han MK, Wang DY, Han B, Zong L, Lan L, et al. A POU3F4 mutation causes nonsyndromic hearing loss in a Chinese X-linked recessive family. Chin Med J. 2017;130:88–92.
Pollak A, Lechowicz U, Kedra A, Stawinski P, Rydzanicz M, Furmanek M, et al. Novel and de novo mutations extend association of POU3F4 with distinct clinical and radiological phenotype of hearing loss. PLoS ONE. 2016;11:e0166618.
Palmer S, Groves N, Schindeler A, Yeoh T, Biben C, Wang CC, et al. The small muscle-specific protein Csl modifies cell shape and promotes myocyte fusion in an insulin-like growth factor 1-dependent manner. J Cell Biol. 2001;153:985–98.
Schindeler A, Lavulo L, Harvey RP. Muscle costameric protein, Chisel/Smpx, associates with focal adhesion complexes and modulates cell spreading in vitro via a Rac1/p38 pathway. Exp Cell Res. 2005;307:367–80.
Acknowledgements
We greatly thank the pedigree members for agreeing to participate in this study. We also appreciate the help and advice of our colleagues. This study was supported by the National Basic Research Program of China (also called 973 Program) (No. 2014CB541702; 2014CB943003), the National Nature Science Foundation of China (No. 81771023; 81470705; 81301172; 81500803), the China Postdoctoral Science Foundation (No. 2017M620359) and the Graduate Innovation Fundation of Central South University (No. 2017ZZTS213).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Deng, Y., Niu, Z., Fan, L. et al. A novel mutation in the SMPX gene associated with X-linked nonsyndromic sensorineural hearing loss in a Chinese family. J Hum Genet 63, 723–730 (2018). https://doi.org/10.1038/s10038-018-0443-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-018-0443-x
This article is cited by
-
Differentiation and functioning of the lateral line organ in zebrafish require Smpx activity
Scientific Reports (2024)
-
Missense mutations in small muscle protein X-linked (SMPX) cause distal myopathy with protein inclusions
Acta Neuropathologica (2021)