Abstract
Several studies reported that autoimmune diseases share a number of susceptibility genes. Of these genes, a SNP rs7708392 in TNIP1 was reported to be associated with systemic lupus erythematosus (SLE). Autoimmune hepatitis (AIH), a rare chronic progressive liver disease, shares some clinical features with SLE. Therefore, we investigated whether the SNP is associated with Japanese AIH. An association study of rs7708392 was conducted in 343 Japanese AIH patients and 828 controls. We found that rs7708392 is associated with AIH (P = 0.0236, odds ratio (OR) 1.26, 95% confidence interval (CI): 1.03–1.54), under the allele model for C allele. Significant differences of clinical characteristics of the AIH patients with or without G allele of rs7708392 were not detected. Of interest, the association was stronger in AIH without HLA-DRB1*04:05 allele (P = 0.0063, Q = 0.0127, OR 1.48, 95% CI: 1.12–1.96), though the association was not detected in AIH with DRB1*04:05. The C allele of rs7708392 was associated with AIH, especially AIH without DRB1*04:05, an already established risk factor.
Similar content being viewed by others
Introduction
Autoimmune hepatitis (AIH) is a very rare chronic progressive liver disease with autoimmune features that mainly affects middle-aged woman [1,2,3,4]. The prevalence of AIH in Japan is 15 per 100,000 people [5]. Type-1 AIH is characterized by elevated serum transaminase and immunoglobulin G levels, the presence of serum anti-nuclear antibodies (ANA), anti-smooth muscle antibodies (ASMA), and interface hepatitis. AIH was originally designated as lupoid hepatitis, because AIH shares some clinical features, the presence of lupus erythematosus cells and ANA, with systemic lupus erythematosus (SLE) [6,7,8]. The etiology of the disease is still unknown, but the genetic and environmental factors are thought to be associated with AIH. AIH is associated with human leukocyte antigen (HLA)-DRB1*03:01 and DRB1*04:01 in European populations [9] and DRB1*04:05 in Japanese populations [10,11,12]. Non-HLA genes might also confer the genetic risk of AIH. A recent genome-wide association study (GWAS) on European type-1 AIH has identified significant genetic risk factors only in HLA region. The GWAS also reported suggestive associations of single nucleotide polymorphisms (SNPs) in SH2B3 and CARD10 genes [13]. However, there are a few reports of genetic association studies of SNPs in non-HLA genes in Japanese AIH; STAT4 [14], CARD10 [15], PTPN22 [16], ICOS [17], and SH2B3 [18].
It was known that autoimmune diseases share a number of susceptibility genes [19]. Associations of SNPs in TNIP1 gene were observed in SLE [20,21,22], psoriasis [23, 24], and systemic sclerosis [25, 26]. Two SNPs, rs7708392 and rs10036748, in TNIP1 gene are in strong linkage disequilibrium in Japanese [22]; rs7708392 is associated with SLE in European [21] and Japanese [22], and rs10036748 in Chinese [20]. TNIP1 (TNFAIP3 interacting protein 1) encodes an adaptor protein binding to A20, an inhibitor of nuclear factor-κB (NF-κB), encoded by TNFAIP3 (tumor necrosis factor-α induced protein 3) gene [27] that is also susceptibility gene for SLE [23, 28]. Because of the clinical similarity between AIH and SLE [7, 8], we carried out a case–control association study of rs7708392 in TNIP1 gene with Japanese AIH and tried to observe the genetic interaction with DRB1 the sole established genetic risk factor confirmed in GWAS.
Materials and methods
Patients and controls
Three hundred forty-three patients with type-1 AIH were enrolled in the register of Japanese National Hospital Organization (NHO) Liver Registry [29, 30]; all the patients without any other types of liver diseases satisfied the criteria of International Autoimmune Hepatitis Group for diagnosis of type I AIH [31]. AIH patients were recruited from NHO hospitals located in various regions of Japan. The healthy controls (n = 828; mean age ± SD, 36.1 ± 10.9, 240 male [29.2%]) included previously reported 513 [22] and newly recruited 67 Japanese healthy individuals recruited in Tokyo and adjacent areas. The DNA samples form 248 Japanese healthy individuals were distributed by the Pharma SNP Consortium (Tokyo, Japan) [32]. All patients and healthy individuals were native Japanese living in Japan. This study was reviewed and approved by the Ethics Committees of NHO central IRB and University of Tsukuba Research Ethics Committee. Written informed consent was obtained from each individual. This study was conducted in accordance with the principles expressed in the Declaration of Helsinki.
Genotyping
Genotyping of rs7708392 [C/G] in TNIP1 gene was performed using TaqMan genotyping assay (Assay ID: C_29349759_10, Thermo Fisher Scientific Inc., Waltham, MA) on 7500 Fast Real-Time PCR System (Thermo Fisher Scientific Inc.) or 7300 Real-Time PCR System (Thermo Fisher Scientific Inc.), according to the manufacturer’s instructions. SNP genotyping results for some of the healthy controls were reported previously [22]. Results of DRB1 genotyping for the AIH patients and the healthy controls were reported previously [12, 33].
Statistical analysis
The distribution of allele frequencies was compared between AIH patients and healthy controls by chi-square analysis using 2×2 contingency tables. The clinical phenotypes of AIH patients without G allele of rs7708392 were compared with those with by χ2 analysis using 2×2 contingency tables or Mann–Whitney’s U Test. Correction for multiple testing was performed by calculating false discovery rate Q value [34]. The sample size of this experiment had the power of 62% to detect association when the relative risk was 1.26 or higher under the allele model (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize) [35].
Results
Demographic features of the type I AIH patients
Table 1 shows the characteristics of the type I AIH patients. Among 343 AIH patients, 303 AIH patients were positive for ANA (88.3%) and 116 were positive for ASMA (38.4%). One hundred seventy seven AIH patients have predisposing DRB1*04:05 allele (51.6%).
Associations of rs7708392 in TNIP1 with AIH
The SNP, rs7708392, in TNIP1 gene was genotyped in Japanese AIH patients and healthy controls. No deviation from Hardy–Weinberg equilibrium was detected in the patients (P = 0.9847) or the controls (P = 0.6889). Allele frequencies in the AIH patients and the healthy controls are shown in Table 2. A significant association was shown for rs7708392 (P = 0.0236, odds ratio (OR) 1.26, 95% confidence interval (CI): 1.03–1.54) under the allele model for C allele.
Differences of clinical features of the AIH patients with or without G allele of rs7708392
We further tested the clinical phenotypes of AIH patients with or without G allele of rs7708392 (Table 3). No significant difference was detected.
Associations of rs7708392 with the AIH subset without DRB1*04:05 allele
Genetic interaction between rs7708392 and DRB1 was tested (Table 4), because HLA is the single genetic risk factor confirmed in GWAS. We found a significant association for rs7708392 with the AIH subset without DRB1*04:05 allele, the already established predisposing allele in the Japanese (P = 0.0063, Q = 0.0127, OR 1.48, 95% CI: 1.12–1.96). However, no difference of allele frequencies was observed between the AIH patients and the healthy controls with DRB*04:05 allele. Thus, rs7708392C allele was associated with AIH without DRB*04:05 allele, though the association was not detected in AIH with DRB1*04:05.
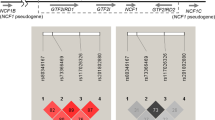
To confirm that rs7708392C and DRB1*04:05 independently contributed to the susceptibility of AIH, conditional logistic regression analysis was conducted (Supplementary Table 1). In unconditioned analysis, rs7708392 (P = 0.0272, OR 1.25, 95% CI: 1.03–1.53) and DRB*04:05 (P = 8.13 × 10−23, OR 3.47, 95% CI: 2.71–4.44) were associated with AIH. In conditioned analysis, the association of rs7708392 remained significant (P = 0.0404, OR 1.25, 95% CI: 1.01–1.54), when conditioned on DRB*04:05. DRB*04:05 was still associated with AIH (P = 1.19 × 10−22, OR 3.46, 95% CI: 2.70–4.44), when conditioned on rs7708392. These data indicated that the two alleles were independently associated with AIH, expected from the different chromosomal location of these loci (chromosome 6 for DRB1 and chromosome 5 for rs7708392).
Discussion
In this study, we found that rs7708392C is a risk allele for type I AIH in a Japanese population. TNIP1 is known to be implicated in the NF-κB signaling pathway [27]. TNIP1 is a shared susceptibility gene in SLE [20,21,22] and AIH, as well as HLA [9,10,11,12, 36] or STAT4 [14, 37], suggesting the presence of the common signaling pathways for immunological reactions in the pathogenesis of these two autoimmune diseases. The involvement of the NF-κB signaling pathway in the pathogenesis of AIH could be explained by the effectivity of corticosteroid treatment on AIH [38]. Because of the common signaling pathways on the pathogenesis between AIH and SLE, the association of rs7708392 would be observed in SLE patients with liver dysfunction and the associations of polymorphisms in TNFAIP3 gene could be detected in AIH patients.
The SNP rs7708392 is located in the intron between exon 1B and exon 2 of TNIP1 gene, but was not thought to be a polymorphism associated with mRNA expression levels of TNIP1 gene [22]. As several splicing forms were reported for TNIP1 gene [27], it is suspected that it may influence splicing isoforms of the TNIP1 gene. Such possibilities will be addressed by comparison of the RNA-seq data of the individuals possessing C/C or G/G genotype. It is alternatively possible that rs7708392 might influence on the expression of the adjacent ANXA6 gene as reported in the eQTL study of whole blood (P = 1.05 × 10−24, http://www.broadinstitute.org/mammals/haploreg/haploreg.php) [39, 40]. Female is predominant in AIH and rs7708392 was reported to influence on the expression of CD74 gene in female [41].
Recent GWAS reported a suggestive association of polymorphisms in non-HLA region, rs3184504 in SH2B3 and rs6000782 in CARD10. However, no polymorphism of rs3184504 was reported in Japanese populations and rs6000782 was not associated with type I AIH in Japanese populations [15]. Thus, based on the similarity between some clinical characteristics of AIH and SLE, the present association study was conducted on the SLE susceptible SNP in Japanese AIH.
We also analyzed the association of rs7708392 after stratifying patients and controls according to DRB1*04:05 allele. A stronger association was detected with DRB1*04:05 negative AIH patients, suggesting the predisposing role of rs7708392 in the absence of the strongest genetic risk factor, DRB1*04:05. The weaker risk factors could be easily detected in the individuals without the strongest risk factor. Otherwise, the weaker risk factors may not contribute to the susceptibility of AIH in individuals with the strongest risk factor. The C allele of rs7708392 was associated with SLE as well as the subsets with renal disorder, but not with the subsets with the age of onset less than 20 years [22]. The SNP in TNIP1 may have some roles in the pathogenesis of some SLE subsets with liver dysfunction and it should be investigated.
As the sample size of this study is limited, the observed association of rs7708392 was modest. The gene–gene interaction between this SNP and DRB1 was barely detected in this modest sample size, because DRB1 is the sole established genetic risk factor confirmed in GWAS. However, other unconfirmed SNPs located outside of the HLA region slightly contributed to the susceptibility of AIH compared to DRB1 [13]. Thus, the analyses of the gene–gene interaction of the SNP with the unconfirmed SNPs were not performed in this study. Although it is quite difficult to increase the sample size of the patients with this rare disease, the association of the SNP with AIH should be confirmed and the gene–gene interaction with the unconfirmed SNPs should be investigated in future independent large-scale studies.
To the best of our knowledge, this is the first report of the association of rs7708392 in TNIP1 with Japanese type I AIH. The associations of rs7708392 should be confirmed in future large-scale studies, though it is difficult to enlarge the sample size of this rare disease.
References
Manns MP, Czaja AJ, Gorham JD, Krawitt EL, Mieli-Vergani G, Vergani D, et al. Diagnosis and management of autoimmune hepatitis. Hepatology. 2010;51:2193–213.
Moy L, Levine J. Autoimmune hepatitis: a classic autoimmune liver disease. Curr Probl Pediatr Adolesc Health Care. 2014;44:341–6.
Carbone M, Neuberger JM. Autoimmune liver disease, autoimmunity and liver transplantation. J Hepatol. 2014;60:210–23.
Wang Q, Yang F, Miao Q, Krawitt EL, Gershwin ME, Ma X. The clinical phenotypes of autoimmune hepatitis: A comprehensive review. J Autoimmun. 2016;66:98–107.
Yoshizawa K, Joshita S, Matsumoto A, Umemura T, Tanaka E, Morita S, et al. Incidence and prevalence of autoimmune hepatitis in the Ueda area, Japan. Hepatol Res. 2016;46:878–83.
Mackay IR, Taft LI, Cowling DC. Lupoid hepatitis. Lancet. 1956;271:1323–6.
Mackay IR, Weiden S, Hasker J. Autoimmune hepatitis. Ann N Y Acad Sci. 1965;124:767–80.
Mackay IR, Taft LI, Cowling DC. Lupoid hepatitis and the hepatic lesions of systemic lupus erythematosus. Lancet. 1959;1:65–69.
Strettell MD, Donaldson PT, Thomson LJ, Santrach PJ, Moore SB, Czaja AJ, et al. Allelic basis for HLA-encoded susceptibility to type 1 autoimmune hepatitis. Gastroenterology. 1997;112:2028–35.
Umemura T, Katsuyama Y, Yoshizawa K, Kimura T, Joshita S, Komatsu M, et al. Human leukocyte antigen class II haplotypes affect clinical characteristics and progression of type 1 autoimmune hepatitis in Japan. PLoS ONE. 2014;9:e100565.
Furumoto Y, Asano T, Sugita T, Abe H, Chuganji Y, Fujiki K, et al. Evaluation of the role of HLA-DR antigens in Japanese type 1 autoimmune hepatitis. BMC Gastroenterol. 2015;15:144.
Maeda Y, Migita K, Higuchi O, Mukaino A, Furukawa H, Komori A, et al. Association between anti-ganglionic nicotinic acetylcholine receptor (gAChR) antibodies and HLA-DRB1 alleles in the Japanese population. PLoS ONE. 2016;11:e0146048.
de Boer YS, van Gerven NM, Zwiers A, Verwer BJ, van Hoek B, van Erpecum KJ, et al. Genome-wide association study identifies variants associated with autoimmune hepatitis type 1. Gastroenterology. 2014;147:443–52 e445.
Migita K, Nakamura M, Abiru S, Jiuchi Y, Nagaoka S, Komori A, et al. Association of STAT4 polymorphisms with susceptibility to type-1 autoimmune hepatitis in the Japanese population. PLoS ONE. 2013;8:e71382.
Migita K, Jiuchi Y, Furukawa H, Nakamura M, Komori A, Yasunami M, et al. Lack of association between the CARD10rs6000782 polymorphism and type 1 autoimmune hepatitis in a Japanese population. BMC Res Notes. 2015;8:777.
Umemura T, Joshita S, Yamazaki T, Komatsu M, Katsuyama Y, Yoshizawa K, et al. Genetic association of PTPN22 polymorphisms with autoimmune hepatitis and primary biliary cholangitis in Japan. Sci Rep. 2016;6:29770.
Higuchi T, Oka S, Furukawa H, Nakamura M, Komori A, Abiru S, et al. Association of a single nucleotide polymorphism upstream of ICOS with Japanese autoimmune hepatitis type 1. J Hum Genet. 2017;62:481–4.
Umemura T, Joshita S, Hamano H, Yoshizawa K, Kawa S, Tanaka E, et al. Association of autoimmune hepatitis with Src homology 2 adaptor protein 3 gene polymorphisms in Japanese patients. J Hum Genet. 2017;62:963–7.
Lettre G, Rioux JD. Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet. 2008;17:R116–121.
Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–7.
Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–33.
Kawasaki A, Ito S, Furukawa H, Hayashi T, Goto D, Matsumoto I, et al. Association of TNFAIP3 interacting protein 1, TNIP1 with systemic lupus erythematosus in a Japanese population: a case–control association study. Arthritis Res Ther. 2010;12:R174.
Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappa B pathways. Nat Genet. 2009;41:199–204.
Zuo X, Sun L, Yin X, Gao J, Sheng Y, Xu J, et al. Whole-exome SNP array identifies 15 new susceptibility loci for psoriasis. Nat Commun. 2015;6:6793.
Allanore Y, Saad M, Dieude P, Avouac J, Distler JH, Amouyel P, et al. Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 2011;7:e1002091.
Bossini-Castillo L, Martin JE, Broen J, Simeon CP, Beretta L, Gorlova OY, et al. Confirmation of TNIP1 but not RHOB and PSORS1C1 as systemic sclerosis risk factors in a large independent replication study. Ann Rheum Dis. 2013;72:602–7.
Verstrepen L, Carpentier I, Verhelst K, Beyaert R. ABINs: A20 binding inhibitors of NF-kappa B and apoptosis signaling. Biochem Pharmacol. 2009;78:105–14.
Kawasaki A, Ito I, Ito S, Hayashi T, Goto D, Matsumoto I, et al. Association of TNFAIP3 polymorphism with susceptibility to systemic lupus erythematosus in a Japanese population. J Biomed Biotechnol. 2010;2010:207578.
Migita K, Watanabe Y, Jiuchi Y, Nakamura Y, Saito A, Yagura M, et al. Evaluation of risk factors for the development of cirrhosis in autoimmune hepatitis: Japanese NHO-AIH prospective study. J Gastroenterol. 2011;46(Suppl 1):56–62.
Migita K, Watanabe Y, Jiuchi Y, Nakamura Y, Saito A, Yagura M, et al. Hepatocellular carcinoma and survival in patients with autoimmune hepatitis (Japanese National Hospital Organization-autoimmune hepatitis prospective study). Liver Int. 2012;32:837–44.
Alvarez F, Berg PA, Bianchi FB, Bianchi L, Burroughs AK, Cancado EL, et al. International Autoimmune Hepatitis Group report: review of criteria for diagnosis of autoimmune hepatitis. J Hepatol. 1999;31:929–38.
Kamatani N, Kawamoto M, Kitamura Y, Harigai M, Okumoto T, Sumino Y. Establishment of B-cell lines derived from 996 Japanese individuals. Tissue Cult Res Commun. 2004;23:71–80.
Oka S, Furukawa H, Kawasaki A, Shimada K, Sugii S, Hashimoto A, et al. Protective effect of the HLA-DRB1*13:02 allele in Japanese rheumatoid arthritis patients. PLoS ONE. 2014;9:e99453.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300.
Dupont WD, Plummer WD Jr. Power and sample size calculations. A review and computer program. Control Clin Trials. 1990;11:116–28.
Furukawa H, Kawasaki A, Oka S, Ito I, Shimada K, Sugii S, et al. Human leukocyte antigens and systemic lupus erythematosus: a protective role for the HLA-DR6 alleles DRB1*13:02 and *14:03. PLoS ONE. 2014;9:e87792.
Kawasaki A, Ito I, Hikami K, Ohashi J, Hayashi T, Goto D, et al. Role of STAT4 polymorphisms in systemic lupus erythematosus in a Japanese population: a case–control association study of the STAT1-STAT4 region. Arthritis Res Ther. 2008;10:R113.
Marx J. How the glucocorticoids suppress immunity. Science. 1995;270:232–3.
Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934.
Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43.
Linden M, Ramirez Sepulveda JI, James T, Thorlacius GE, Brauner S, Gomez-Cabrero D, et al. Sex influences eQTL effects of SLE and Sjogren’s syndrome-associated genetic polymorphisms. Biol Sex Differ. 2017;8:34.
Acknowledgements
We would like to acknowledge NHO-AIH study group. The members of the NHO-AIH study group are: Kiyoshi Migita (Department of Rheumatology, School of Medicine, Fukushima Medical University), Masashi Ohtani, Seigo Abiru, Yuka Jiuchi, Shinya Nagaoka, Sung Kwan Bae, Atsumasa Komori, Masashi Ohtani, Satoru Hashimoto, Shigemune Bekki, Katsumi Yamasaki, Hiroshi Yatsuhashi, Hiromi Ishibashi (NHO Nagasaki Medical Center), Minoru Nakamura (Department of Hepatology, Nagasaki University Graduate School of Biomedical Sciences), Michio Yasunami (Life Science Institute, Saga-ken Medical Centre Koseikan), Yukio Watanabe, Yoko Nakamura (NHO Sagamihara National Hospital), Michiyasu Yagura (NHO Tokyo National Hospital), Tatsuji Komatsu (NHO Yokohama Medical Center), Masaaki Shimada (NHO Nagoya Medical Center), Hiroshi Kouno (NHO Kure Medical Center), Taizo Hijioka (NHO Osaka Minami Medical Center), Motoyuki Kohjima (NHO Kyushu Medical Center), Michio Kato (NHO Minami Wakayama Medical Center), Kaname Yoshizawa (NHO Shinshu Ueda Medical Center), Hajime Ohta (NHO Kanazawa Medical Center), Eiichi Takezaki (NHO Higashi Hiroshima Medical Center), Hideo Nishimura (NHO Asahikawa Medical Center), Takeaki Sato (NHO Kokura Medical Center), Keisuke Ario (NHO Ureshino Medical Center), Noboru Hirashima (NHO Higashi Nagoya National Hospital), Yukio Oohara (NHO Hokkaido Medical Center), Haruhiro Yamashita (NHO Okayama Medical Center), Atsushi Naganuma (NHO Takasaki General Medical Center), Toyokichi Muro (NHO Oita Medical Center), Hironori Sakai (NHO Beppu Medical Center), Eiji Mita (NHO Osaka Medical Center), Kazuhiro Sugi (NHO Kumamoto Medical Center), Fujio Makita (NHO Nishigunma National Hospital).
Funding
The work was supported by Grants-in-Aid for Clinical Research from National Hospital Organization. The funders had no role in study design, data collection and analysis, decision to publish, or preparing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Oka, S., Higuchi, T., Furukawa, H. et al. Association of a single nucleotide polymorphism in TNIP1 with type-1 autoimmune hepatitis in the Japanese population. J Hum Genet 63, 739–744 (2018). https://doi.org/10.1038/s10038-018-0440-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-018-0440-0