Abstract
Biallelic mutations of the gene encoding diphthamide biosynthesis 1 (DPH1, NM_001383.3) cause developmental delay, dysmorphic features, sparse hair, and short stature (MIM *603527). Only two missense DPH1 mutations have been reported to date. Here, we describe a consanguineous family with two siblings both showing developmental delay, agenesis of the corpus callosum, dysmorphic facial features, sparse hair, brachycephaly, and short stature. By wholeexome sequencing, a homozygous frameshift mutation in DPH1 (c.1227delG, p.[Ala411Argfs*91]) was identified, which is likely responsible for the familial condition. The unique clinical features of the affected siblings are cleft palate and absent renal findings.
Similar content being viewed by others
Introduction
DPH1 encodes a component of a multi-protein complex—formed together with DPH2, DPH3, DPH4, and DPH5—for diphthamide biosynthesis [1, 2]. To date, at least two DPH1 mutations have been reported, which cause intellectual disability and dysmorphic features [3, 4]. However, the pathogenetic mechanisms by which the DPH1 mutations produce the clinical phenotype are unknown. Here, we report the identification of a consanguineous family with two siblings both showing dysmorphic features and developmental delay caused by a novel homozygous DPH1 mutation, identified by wholeexome sequencing (WES). We also discuss how the genetic mutation might contribute to the clinical features in this family.
Materials and methods
Samples
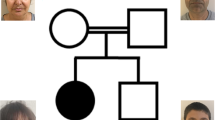
Peripheral blood samples were collected from two affected siblings and their consanguineous parents (Fig. 1a) after a written informed consent was obtained. Genomic DNA was extracted from the peripheral blood leukocytes with a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The study protocol was approved by the institutional review board of Yokohama City University School of Medicine.
Genetic analysis of the pedigree. a Pedigree. The left half of each symbol represents phenotypes associated with DPH1 mutations, and the right half shows craniosynostosis caused by the FREM1 mutation. b Brain CT of Patient 1 at the age of 4 years. Cephalic index is calculated as 85.6%. Possible agenesis of the corpus callosum is seen. c Chromatograms of DPH1 (left) and FREM1 mutations (right). d Schematic illustration of DPH1 cDNA (upper panel) and DPH1 protein (lower panel). The red box shows the diphthamide synthase domain. e Schematic illustration of FREM1 cDNA (upper panel) and FREM1 protein (lower panel). Boxes within the arrows show the following domains: blue, transmembrane region; yellow, cadherin-3; orange, calx-beta; green, C-type lectin or carbohydrate-recognition domain. All functional domains were predicted by SMART (http://smart.embl-heidelberg.de/smart/). Previously reported mutations are shown in black and mutations detected in this study are shown in red (c, d). WT wild type, mut mutant allele, DPH1 diphthamide biosynthesis 1, FREM1 FRAS1-related extracellular matrix 1
Genetic analysis
WES was performed as reported previously [5]. In brief, 3 μg of genomic DNA was sheared and captured using a SureSelect Human All Exon V5 kit (Agilent Technologies, Santa Clara, CA) and sequenced on a HiSeq2000 system (Illumina, San Diego, CA) with 101-bp paired-end reads. Based on the autosomal recessive model, variants with minor allele frequencies of >0.005 in the Exome Aggregation Consortium (ExAC), the National Heart Lung and Blood Institute Exome Sequencing Project (NHLBI-ESP 6500), or Human Genetic Variation Database (HGVD) and variants that were found in the in-house exome data of five or more individuals (from a total of 575 Japanese subjects) were excluded from further analysis. In addition, variants were filtered based on an autosomal dominant model; variants with allele frequencies of >0 in any of the public databases listed above and in-house data were excluded (Supplemental Table 1). Variants on chromosome X were also filtered based on an X-linked recessive model (Supplemental Table 1). Non-excluded variants were further validated by Sanger sequencing on an ABI 3500 Genetic analyzer (Applied Biosystems, Foster City, CA) and analyzed with Sequencher software (Gene Codes, Madison, WI). Homozygous regions were investigated using HomozygosityMapper (http://www.homozygositymapper.org/) [6].
Results
Proband (II-2) (Fig. 1a) was a 10-year-old Iranian boy born as the second child to healthy consanguineous parents. His elder brother had hypotonia, heart disease, and gastroesophageal reflux. He died at 6 months, and the cause of death was recorded as sudden infant death syndrome. In the proband, brachycephaly and a cleft palate were noticed at birth, and agenesis of the corpus callosum (ACC) and cerebellar vermis hypoplasia were found by an MRI at the age of 1 year. His brain CT at 4 years showed brachycephaly (cephalic index of 85.6%) as well as possible ACC (Fig. 1b). He presented with sparse and thin hair, distinctive features (widely spaced eyes with scleral pigmentation, downslanted palpebral fissures, epicanthal folds, depressed nasal bridge, low-set ears, and widely spaced teeth), a bilateral single simian crease, short stature (120 cm, −3 SD), and kyphoscoliosis. His IQ was 35–49 at the age of 10 years. The first seizure (tonic, generalized) occurred at 5 months, and phenobarbital and sodium valproate were prescribed until the age of 8 years. His karyotype was normal. His kidneys were normal by ultrasound examination.
His 4-year-old sister (II-3) (Fig. 1a) showed similar clinical features (developmental delay, brachycephaly, sparse hair and eyebrows, widely spaced eyes with scleral pigmentation, downslanted palpebral fissures, epicanthal folds, depressed nasal bridge, low-set ears, widely spaced teeth, and short stature) with some differences (details are described in Table 1). She did not exhibit kyphoscoliosis or seizures, but had an atrial septal defect (ASD), which was surgically repaired at 16 months of age. A recent echocardiogram revealed mild to moderate ASD, residual patent ductus arteriosus with trivial shunt, and mildly dilated right atrium and ventricle. Brain MRI identified ACC and a cystic posterior fossa at 2 years of age. Her karyotype was normal. Her kidneys were normal by ultrasound examination.
To identify the genetic cause of the disease, we performed WES on proband II-2. We first considered the autosomal recessive model and found a homozygous frameshift mutation in DPH1, NM_001383.3: c.1227delG, p.(Ala411Argfs*91), which was confirmed in both affected siblings by Sanger sequencing (Fig. 1c, d). Both parents were heterozygous carriers of the mutation. Homozygosity mapping in the proband (II-2) showed a 0.5-Mb homozygous stretch (chromosome 17: 1,761,593–2,297,571 bp), including DPH1.
We also examined other pathogenic variants associated with the clinical features regardless of the inheritance models. Sanger sequencing identified a heterozygous nonsense mutation in FREM1, NM_144966.5: c.4312C>T, p.(Arg1438*) in both affected siblings and their father (Fig. 1c, e). No other variants were detected.
Discussion
We identified a novel homozygous mutation in DPH1: c.1227delG, p.(Ala411Argfs*91). To date, only two DPH1 mutations in eight individuals from four families have been reported: c.17T > A p.(Met6Lys) and c.701T > C, p.(Leu234Pro) [3, 4, 7]. Ten individuals, including the two siblings from the present study, with biallelic DPH1 mutations show cardinal clinical features including abnormal skull shape (trigonocephaly, scaphocephaly, or prominent forehead accompanied with metopic ridge), distinctive face (downslanted palpebral fissures, low set ears, depressed nasal bridge, and sparse hair on the scalp, eyelashes, and/or eyebrows), short stature, developmental delay, and intellectual disability (Table 1) [3, 4, 7]. Heart and brain malformations are also frequently observed in individuals with DPH1 mutations. Cardiac and brain anomalies are commonly observed in four individuals with p.Leu234Pro and the current affecting siblings with p.Ala411Argfs*91, but not in individuals with p.Met6Lys (Table 1) [3, 4, 7]. Cranial abnormalities are consistently found in all individuals with biallelic DPH1 mutations, including scaphocephaly and sagittal craniosynostosis (p.Met6Lys) [3], trigonocephaly (p.Leu234Pro) [4, 7], and brachycephaly (p.Ala411Argfs*91 in the current family). Epilepsy in two individuals, including our case (II-2), responded well to treatment [3]. Cleft palates are unique features and no renal diseases are noted in our family (Table 1).
Dph1 knockout mice exhibit developmental and growth delay, immature lung, cleft palate, abnormal neural tube formation in the midbrain, and perinatal or prenatal death [8, 9]. Conditional ablation of Dph1 in neural crest cells results in craniofacial defects with hypoplastic nasal bone, lower jaw, and cleft palate. [8] Cleft palate in Dph1 knockout mice drew our attention, as it is a unique feature in the current family (the homozygous truncation mutation in the family is likely a knockout).
We also found a heterozygous nonsense mutation in FREM1 in both affected siblings and their father. FREM1 encodes an extracellular matrix protein that plays an important role in epidermal differentiation and fusion of midline structures during craniofacial development [10]. Biallelic FREM1 mutations cause Manitoba oculotrichoanal (MOTA) syndrome (MIM #248450) or bifid nose with or without anorectal and renal anomalies (BNAR syndrome; MIM #608980). Heterozygous FREM1 mutations cause trigonocephaly (MIM #614485) with variable severity and incomplete penetrance [11]. Similarly, heterozygous Frem1 knockout mice show altered cranial frontal bone curvature and have a milder phenotype than homozygous mutant mice [11]. Retrospectively, the father’s cranial bones were carefully examined, and only partial synostosis of the metopic suture was recognized. Therefore, there is a possibility that FREM1 may contribute brachycephaly in the siblings. However, since the other phenotype has not been reported in the mutation of FREM1, the single mutated gene (DPH1) is able to explain the cause of the all phenotype of the two siblings.
In conclusion, we found a novel homozygous DPH1 mutation in two affected siblings, which is likely causative for the clinical findings. Only three DPH1 mutations have been described to date, including the current one. Our family provides insight into genotype–phenotype correlations. However, further studies of families with DPH1 mutations are needed to clarify how the genotype translates into phenotype.
References
Chen CM, Behringer RR. Cloning, structure, and expression of the mouse Ovca1 gene. Biochem Biophys Res Commun. 2001;286:1019–26.
Nobukuni Y, Kohno K, Miyagawa K. Gene trap mutagenesis-based forward genetic approach reveals that the tumor suppressor OVCA1 is a component of the biosynthetic pathway of diphthamide on elongation factor 2. J Biol Chem. 2005;280:10572–7.
Loucks CM, Parboosingh JS, Shaheen R, Bernier FP, McLeod DR, Seidahmed MZ, et al. Matching two independent cohorts validates DPH1 as a gene responsible for autosomal recessive intellectual disability with short stature, craniofacial, and ectodermal anomalies. Hum Mutat. 2015;36:1015–9.
Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, et al. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10:148–61.
Miyake N, Tsukaguchi H, Koshimizu E, Shono A, Matsunaga S, Shiina M, et al. Biallelic mutations in nuclear pore complex subunit NUP107 cause early-childhood-onset steroid-resistant nephrotic syndrome. Am J Hum Genet. 2015;97:555–66.
Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper—an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–9.
Seidahmed MZ, Alkuraya FS, Shaheed M, Al Zahrani M, Al Manea W, Mansour F, et al. Ritscher-Schinzel (cranio-cerebello-cardiac, 3C) syndrome: report of four new cases with renal involvement. Am J Med Genet A. 2011;155A:1393–7.
Yu YR, You LR, Yan YT, Chen CM. Role of OVCA1/DPH1 in craniofacial abnormalities of Miller-Dieker syndrome. Hum Mol Genet. 2014;23:5579–96.
Chen CM, Behringer RR. Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev. 2004;18:320–32.
Alazami AM, Shaheen R, Alzahrani F, Snape K, Saggar A, Brinkmann B, et al. FREM1 mutations cause bifid nose, renal agenesis, and anorectal malformations syndrome. Am J Hum Genet. 2009;85:414–8.
Vissers LE, Cox TC, Maga AM, Short KM, Wiradjaja F, Janssen IM, et al. Heterozygous mutations of FREM1 are associated with an increased risk of isolated metopic craniosynostosis in humans and mice. PLoS Genet. 2011;7:e1002278.
Acknowledgements
We thank the patient and his family for participating in this work. This work was supported by grants from Research on Measures for Intractable Diseases, Comprehensive Research on Disability Health and Welfare, the Strategic Research Program for Brain Science, the Initiative on Rare and Undiagnosed Diseases in Pediatrics, and the Initiative on Rare and Undiagnosed Diseases in Adults from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Grants-in-Aid for Scientific Research (A, B, and C) from the Japan Society for the Promotion of Science; the fund for the Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program in the Project for Developing Innovation Systems from the Japan Science and Technology Agency; grants from the Ministry of Health, Labour and Welfare; the Takeda Science Foundation; and the Ichiro Kanehara Foundation for the Promotion of Medical Science & Medical Care. We thank Edanz Group (www.edanzediting.com) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Sekiguchi, F., Nasiri, J., Sedghi, M. et al. A novel homozygous DPH1 mutation causes intellectual disability and unique craniofacial features. J Hum Genet 63, 487–491 (2018). https://doi.org/10.1038/s10038-017-0404-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-017-0404-9
This article is cited by
-
DPH1 syndrome: two novel variants and structural and functional analyses of seven missense variants identified in syndromic patients
European Journal of Human Genetics (2020)
-
Hemorrhagic stroke and renovascular hypertension with Grange syndrome arising from a novel pathogenic variant in YY1AP1
Journal of Human Genetics (2019)
-
Genetic abnormalities in a large cohort of Coffin–Siris syndrome patients
Journal of Human Genetics (2019)