Abstract
The Mongol Empire had a significant role in shaping the landscape of modern populations. Many populations living in Eurasia may have been the product of population mixture between ancient Mongolians and natives following the expansion of Mongol Empire. Geneticists have found that most of these populations carried the Y-haplogroup C3* (C-M217). To trace the history of haplogroup (Hg) C3* and to further understand the origin and development of Mongolians, ancient human remains from the Jinggouzi, Chenwugou and Gangga archaeological sites, which belonged to the Donghu, Xianbei and Shiwei, respectively, were analysed. Our results show that nine of the eleven males of the Gangga site, two of the eight males of Chengwugou site and all of the twelve males of Jinggouzi site were found to have mutations at M130 (Hg C), M217 (Hg C3), L1373 (C2b, ISOGG2015), with the absence of mutations at M93 (Hg C3a), P39 (Hg C3b), M48 (Hg C3c), M407 (Hg C3d) and P62 (Hg C3f). These samples were attributed to the Y-chromosome Hg C3* (Hg C2b, ISOGG2015), and most of them were further typed as Hg C2b1a based on the mutation at F3918. Finally, we inferred that the Y-chromosome Hg C3*-F3918 can trace its origins to the Donghu ancient nomadic group.
Similar content being viewed by others
Introduction
The Mongol Empire, built by Genghis Khan ~800 years ago, was the largest contiguous land empire in world history [1]. It covered most of Eurasia including all of modern-day China, Korea, the Caucasus, Central Asia and substantial portions of modern Eastern Europe [2]. The expansion of the Mongol Empire resulted in the extensive migration of populations. It is believed that the Mongol Empire had a significant role in shaping the landscape of modern populations who are living in the area ever covered by the Mongol Empire [3, 4]. This fact attracted geneticists to investigate the Mongolians and their related populations. Studies have shown that haplogroup (Hg) C3-M217 was predominant in the Mongolians and Central Asians who are regarded as the descendants of the Ancient Mongolians, with the frequencies ranging from 30 to 90% [5]. Moreover, the Hg C3*, assigned according to haplotypes that were outside of the single nucleotide polymorphisms (SNP)-defined subgroups C3a-M93, C3b-P39, C3c-M48, C3d-M407, C3e-P53.1 and C3f-P62, was observed at a high frequency in Kereys of Kazakhstan, Mongolians, Hazaras of Pakistan (the highest frequencies reached 89.3%), Kazaks from Xingjiang of China and Mongolians from inner Mongolia and Heilongjiang of China, who are related to ancient Mongolians in terms of their origins or inhabitations [5, 6]. Many geneticists suggested that this lineage might originate from Mongolia [5, 7], and they even discovered that the Hg C3* star-cluster, identified according to 17 STR loci, might be attributed to Genghis Khan [7, 8]. As such, the Hg C3* could be an important genetic marker to track the lineage of the Mongol Empire. Therefore, tracing the history of Hg C3* is of great help to understand the origin and development of the Mongolians.
Most historians have concluded that the Mongolians can trace their origin to the Donghu nomadic tribes [9]. The Donghu formed as early as the fourth century bc and lived in the eastern area of Xiongnu [10]. They were flourishing in North China for many years according to some accounts found in ancient Chinese literature [10]. Around 206 bc, the Donghu were defeated by the Xiongnu, and the remaining members of this group survived as the Xianbei and Wuhuan, living in Northeast China for hundreds of years. Around 89 ad, the Xianbei allied with the Han Empire to disperse the Xiongnu and as a result became powerful and highly populated, taking almost all of the territory previously held by the Xiongnu. Around 5 ad, parts of the Xianbei population gradually developed into the Shiwei ancient nomadic group. During the tenth century, the Shiwei population began to migrate westward, and migrated into the region currently known as Mongolia [11]. They developed into the Mongolians and built one of the greatest empires in history; they controlled ~22% of the world’s total land area at their peak [12].
Recently, three archaeological sites, Jinggouzi (JGZ), Chenwugou (CWG) and Gangga (GG), were found in the inner Mongolia region of China. The Jinggouzi site was excavated in the sloping loess hillock of a wadi (118.14°E, 43.23°N) in the northern bank of the upper Xilamulun River in Chifeng City of the inner Mongolia region of China [13] (Fig. 1). Many animal bones, saddleries, bows and arrows, but no farming tools or farming products, were discovered at this archaeological site (Fig. 1a) [13]. The radiocarbon (14C) measurements (2485 ± 45 bp) showed that the Jinggouzi site was estimated to have been used in the late Spring–Autumn period (770–476 bc) and the early War States period (475–221 bc) [13]. This timing indicates that the Jinggouzi site can be associated with the Donghu culture [13]. The Chenwugou site is located in Huade County (114.19°E, 42.04°N) in Ulanqab city of the inner Mongolia region of China (Fig. 1). On the basis of the funerary objects and burial customs discovered at this site (Fig. 1b), archaeologists suggested that these remains could be dated to the fourth and fifth centuries ad and most likely belong to the tribe of the Xianbei alliance [14]. The Gangga site is located in Chen Barag Banner (119.45°E, 49.33°N) of the inner Mongolia region of Northeast China (Fig. 1) [15]. The radiocarbon analysis of the remains from the Gangga site dated these samples to the eighth to tenth centuries ad. Several items were discovered in Gangga burial sites, including as pottery, wooden saddles, birth bark quivers, iron arrowheads, bronze belt ornaments and agate beads (Fig. 1c). These findings, in conjunction with evidence of certain mortuary practices, indicated that the Shiwei population resided in the Gangga region [15].
Geographic location of the Jinggouzi, Chenwugou and Gangga sites. The grey area marked on the map represents the territory of the Mongol Empire. The region on the upper right of the graph shows the ancestors of the Mongolians at different historical periods. a The human remains and funerary objects excavated from the Jinggouzi site. b The human remains and funerary objects excavated from the Chenwugou site. c The human remains and funerary objects excavated from the Gangga site
In this study, we aimed to determine the genetic variants of the Y-chromosome in human remains excavated from three archaeological sites through analysis of SNP. This analysis can provide important information concerning the genetic structure of the ancestors of the Mongolians, which could lead to a better understanding of the framework of the ancient Mongolian population.
Materials and methods
Sample selection
Human remains from 12, 12 and 16 individuals were selected from the Jinggouzi, Chenwugou and Gangga sites, respectively. Skulls and pelvic bone samples from all subjects were used for anthropological sex identification, and teeth were used for molecular biological analysis. All human remains used for this study were excavated by the Institute of Archaeology of Chinese Academy of Social Sciences and Institute of Cultural Relics and Archaeology of inner Mongolia with permission from the State Administration of Cultural Heritage of China.
Methods to avoid DNA contamination and monitor authenticity
Several precautions were taken to prevent modern human contamination. Researchers in the pre-PCR lab wore full-body protective clothing, facemasks and several layers of gloves. They also followed strict workflow protocols when doing lab work. The post and pre-PCR labs are located in separate buildings 2 km apart. Sample preparation, DNA extraction and purification, and PCR setup were completed in the pre-PCR lab. Every step in the pre-PCR lab was performed in hoods that were irradiated with UV for at least 30 min and cleaned with DNA-Off (MPbio, USA) before, between, and after each use. Rooms and other equipment were also treated as described above. No other molecular biology tests were performed in this building. PCR and sequencing were carried out in the post-PCR lab. All the experiments were performed in parallel using duplicate teeth from each individual to check for reproducibility. The HVR1 and Y-SNP of all researchers who worked in the lab were sequenced to check for contamination.
DNA extraction
Teeth were soaked in a 5% sodium hypochlorite solution for 20 min, rinsed once with ddH2O and 95% ethanol, and then dried on each side in a UV-irradiation box. Overall, 200 mg of dentin per sample was ground into a fine powder using a dental drill (STRONG 90), and digested in 5 mL of EDTA (0.5 mol, pH 8) and 70 μL of proteinase K (100 mg/mL) for 12–16 h in a rotating hybridisation oven at 55 °C. After centrifuging the samples at 7500 rpm for 5 min, the supernatant was condensed to ~100 μL at 6300 rpm using ultrafiltration tubes (Centricon® YM-10). DNA was then extracted and eluted to a final volume of 70 μL using the QIAquick PCR Purification Kit (Qiagen, Germany) according to the manufacturer’s protocol.
Sex identification and Y-chromosome SNP typing
For sex determination, the amelogenin gene fragment was amplified, and samples detected as male were further studied. To identify the Y-chromosome haplogroups of the samples, SNP markers defining the whole Hg C (M216) and its subgroups C1 (M8), C2 (M38), C3 (M217) and C5 (M356) were assayed according to ISOGG2011. The SNPs M93, P39, M48, M407 and P62 were further typed to detect the haplogroups: C3a, C3b, C3c, C3d and C3f. The markers L1373 (C2b), F3918 (C2b1a), F2613 (C2e) and F978 (C2f), which belong to the ISOGG2015 Y DNA haplogroup tree, were also selected to identify whether the samples could be attributed to Hg C3e. All primers are listed in Table 1.
Results
All of the 12 JGZ samples revealed reproducibly successful amplifications of the Y-chromosome SNP analysis, and they contained the same mutations as follows: a C → T transition at M130 defined these samples as Hg C; the presence of A → C transition at M217 (Hg C3), and the absence of mutations at M8 (Hg C1), M38 (Hg C2), M356 (Hg C5) typed these samples as Hg C3 (Hg C4 defined by the marker M347 and Hg C6 defined by the marker P55 were not considered in this study because they are completely restricted to Australia and New Guinea, respectively [16, 17]); the absence of mutations at M93 (Hg C3a), P39 (Hg C3b), M48 (Hg C3c, M407 (Hg C3d) and P62 (Hg C3f) further attributed them to Hg C3* or C3e. Next, these samples showed mutations at L1373 and F3918 and were attributed to the Hg C2b1a of ISOGG2015 Y DNA haplogroup tree, which should be the same as the Hg C3* of ISOGG2011. Again, the SNP P53.1 was used to identify the Hg C3e of ISOGG2011 to the Hg C2c of ISOGG2015 (Fig. 2). Thus, these individuals can be typed as Hg C3* but not Hg C3e. From a total of 12 CWG individuals analysed, 8 individuals revealed reproducibly successful amplifications of molecular sex determination using amelogenin analysis. Only two individuals (CWG9 and CWG11) yielded an authentic result after the Y-chromosome SNP analysis. Like the samples from JGZ, both of them contained the mutations at M130, M217 and L1373, with the absence of the mutations at M93, P39, M48, M407 and P62. Thus, two individuals were also classified into the Hg C3*. Eleven of 16 individuals from GG were classified as male through sex identification using the amelogenin gene. Nine out of the 11 males also expressed the Y-haplogroup C-M130 and C-M217, and did not show mutations at M93, P39, M77, M407 and P62. Again, they showed the mutations at C-L1373 (C2b, ISOGG2015) and were further categorised into the sub-haplogroup F3918 (C2b1a), except for GG8 and GG20 because these two individuals failed in this specific typing. Finally, these nine samples were attributed to Hg C3*. The Y-chromosome results from these ancient samples from three sites are shown in Table 2.
Discussion
Authenticity of results
Throughout this study, no amplification products of the Y-chromosome SNP analysis could be obtained from any of the negative controls, including female human remains. The molecular sex identification results were inconsistent with morphological sex assignments, and the Y-chromosome SNP analysis yielded consistent results through three or more independent extractions. None of the male researchers shared the Hg C3* classification of the samples used in this study. Finally, we considered that 9 of 16 GG samples and 2 of 12 CWG samples yielded authentic results. For the JGZ samples, we obtained the Y-chromosome SNP analysis results. The main reason was that the JGZ samples had been analysed in previous studies, and the Y-chromosome SNP analysis was performed directly in 12 individuals identified as male. In fact, 42 individuals had been analysed in a previous study, and 12 individuals showed the mutations at M130 (Hg C), M217 (Hg C3) and P53.1 (Hg C3e) but not at M93 (Hg C3a), P39 (Hg C3b), M77 (Hg C3c), M407 (Hg C3d) and P62 (Hg C3f). We considered that these samples should be classified as Hg C3e [18]. However, we recently found using our primers that the fragment containing the P53.1 site almost completely matched a fragment on Chromosome X when using the NCBI BLAST tool, and the female sample also yielded a positive result (Fig. 3). We cannot determine whether or not the samples should be attributed to Hg C3e or Hg C3*. Thus, the 12 JGZ samples of teeth, which were determined to be males and were amplified successfully in the Y-chromosome SNP analysis, were analysed again in this study. We identified more SNP loci to answer this question. Finally, the mutations at L1373 and F3918 were found in the JGZ samples. According to ISOGG2015, Hg C2 and Hg C2b1a were identified based on the presence of the mutations at L1373 and F3918, respectively, whereas the sample could be attributed to Hg C2c if it showed the mutation at P53.1 (Fig. 2). Given that the JGZ samples did not contain the mutation at P53.1, these samples were typed as C3e and could be the result of amplification in chromosome X (Fig. 3). Together with the absence of mutations at M93, P39, M48, M407 and P62, we deduced that the JGZ ancient people can be attributed to C3*.
The history of haplogroup C3*
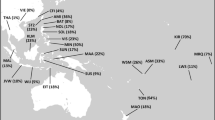
Since Genghis Khan’s era, the Mongolian population, including the Gold Family, invaded neighbouring lands outward from the Mongolian plateau, resulting in gene flow from the Mongolian plateau to most of Eurasia [9]. Thus, many populations living in areas that stretch from the Sea of Japan on the East to the Mediterranean on the West and Siberia on the North to the Persian Gulf on the South, are regarded as the descendants of the Mongolians. For example, Kerey, which is the dominant clan in Kazakhstan, can trace their origins to the Mongolians. This group showed the highest frequency (76.5%) of individuals carrying the Hg C3* [8] (Fig. 4). Hg C3* is also found at high frequencies among Central Asian peoples and indigenous Siberians, such as the Buryats, Evens, Evenks, Kazakhs, Mongolians and Udegeys [5], who are regarded as the descendants of ancient Mongolians. As such, Hg C3* is an important paternal lineage from Mongolia.
The development of Mongolia and the frequencies of haplogroup C3* in modern Eurasians. a The development of Mongolia. b The frequencies of haplogroup C3 in modern Eurasians. The dotted line represents the approximate boundary between the Xiongnu and the Donghu. The black and grey arrows denote the migration of the Donghu and Mongolians, respectively
However, Hg C3* is the ancestral state at the M93, P39, M48, M407, P53.1 and P62 loci, and includes haplotypes that are outside of Hg C3a-f. It is not a monophyletic group and has several sub-haplogroups including Hg C3* star-cluster and other sub-branches. Many geneticists realised that Hg C3* star-cluster, currently referred to as C3-F1918 [19], was the paternal lineage of Genghis Khan or his close relatives, and they suggested that this lineage might have originated in Mongolia about 1000 years ago [7]. There is no denying that the majority of C3* haplogroups in previous studies belong to the C3* star-cluster, while in this study, most of the samples (about 78%) showed the mutation at F3918. Obviously, they do not belong to the Hg C3* star-cluster, the well-known lineage and the majority of C3*-M217 haplogroups, according to the revised phylogenetic tree for haplogroup C3*-DYS448del [19]. However, it does not mean that the haplogroup carried by the samples in this study is not related to the ancient Mongolians and even with Genghis Khan. First, untill now, the paternal lineages of Genghis Khan and the Gold Family are unclear. No enough evidence supports that Hg C3* is directly related to Genghis Khan because all members of the imperial family of the Mongol Empire were buried without identifying signs, and the burial site of Genghis Khan has never been found. Recently, the study of three ancient individuals who were thought to belong to the Gold Family proposed that Genghis Khan and his family carried Hg R1b-M343 [20]. Second, ancient Mongolian populations, including the Gold Family, comprises several clans; Genghis Khan, for example, belonged to the Borjigin clan [9]. These clans should belong to different Y-haplogroups. In recent study, Hg C3*-F1756, as the sub-branch of Hg C3*-F3918, was also found in Mongolic-speaking populations (4–12%) [19]. Interestingly, 9 of the 20 graves from the GG site were furnished with log coffins. This burial custom was also performed by the royal families of the ancient Mongolians, aptly named the Gold Family. In fact, only members of the Gold Family were allowed to be buried with log coffins [15]. This implies that these 9 GG individuals might be the nobility of the Shiwei population, as well as the ancestors of the royal family of ancient Mongolia. All of the GG individuals who were buried in the traditional log coffins were attributed to the Y-chromosome Hg C3*-L1373 in this study, and seven of them were typed exactly as Hg C3*-F3918. This means that Shiwei population and the royal family of ancient Mongolia should carry Hg C3*-F3918. According to historical documents, the Shiwei population was formed from segments of the Xianbei populations. In this study, CWG samples from the Xianbei nomadic group, according to archaeological studies, were chosen. Although only two samples yielded results in this study, both of them were calculated as Hg C3*-L1373. Although we cannot ensure the possibility that they belong to Hg C3*-F3918, it can at least be concluded that the Xianbei nomadic group included Hg C3*. Xianbei was one of two major tribe alliances resulting from the division of the Donghu. The Donghu was one of the ancient nomadic populations in North China [10], and the people are considered to be the earliest ancestors of the Mongolians [9]. In this study, we found that all the males of the JGZ people, who belonged to the Donghu population, were typed into Hg C3*-L1373, and 92% of them were further attributed to Hg C3*-F3918. This indicates that Hg C3*-F3918 is one of the Y-chromosome lineage markers of the Donghu population and originated as early as 2500 years ago.
Overall, three ancient populations analysed in this study share similar Y-chromosome haplogroup C3*-L1373 and 78% of the samples were attributed to Hg C3*-F3918. Moreover, many modern populations who trace their origin to Mongolia also belong to this haplogroup. This finding indicates genetic continuity of Hg C3*-F3918 from the Donghu nomadic group to the Shiwei ancient people, and finally to modern Mongolians and their related populations. Thus, we infer that the Y-chromosome Hg C3*-F3918 can be traced to the Donghu ancient nomadic group from ~2500 years ago.
References
Morgan D. The Mongols. New Jersey: Wiley-Blackwell; 2007.
Bentley JH. Old world encounters: cross-cultural contacts and exchange in pre-modern times. New York, NY: Oxford University; 1993.
Balaresque P, Poulet N, Cussat-Blanc S, Gerard P, Quintana-Murci L, Heyer E, et al. Y-chromosome descent clusters and male differential reproductive success: young lineage expansions dominate Asian pastoral nomadic populations. Eur J Hum Genet. 2015;23:1413–22.
Bai H, Guo X, Zhang D, Narisu N, Bu J, Jirimutu J, et al. The genome of a Mongolian individual reveals the genetic imprints of Mongolians on modern human populations. Genome Biol Evol. 2014;6:3122–36.
Zhong H, Shi H, Qi XB, Xiao CJ, Jin L, Ma RZ, et al. Global distribution of Y-chromosome haplogroup C reveals the prehistoric migration routes of African exodus and early settlement in East Asia. J Hum Genet. 2010;55:428–35.
Derenko MV, Maliarchuk BA, Wozniak M, Denisova GA, Dambueva IK, Dorzhu CM, et al. Distribution of the male lineages of Genghis Khan’s descendants in northern Eurasian populations. Genetika. 2007;43:422–6.
Zerjal T, Xue Y, Bertorelle G, Wells RS, Bao W, Zhu S, et al. The genetic legacy of the Mongols. Am J Hum Genet. 2003;72:717–21.
Abilev S, Malyarchuk B, Derenko M, Wozniak M, Grzybowski T, Zakharov I. The Y-chromosome C3* star-cluster attributed to Genghis Khan’s descendants is present at high frequency in the Kerey clan from Kazakhstan. Hum Biol. 2012;84:79–89.
Du R, Yip VF. Ethnic groups in China. Beijing: Science Press; 1993.
Lin G. A History of Donghu. Hohhot, Inner Mongolia: Inner Mongolia People’s Publishing House; 1989.
Sun J. The origin of Shiwei. Seeking Truth. 1985;1:88–94.
http://my.raex.com/~obsidian/earthrul.html. Accessed 20 June 2017.
Xin Wl, la T, Gang ZY. Lin Xi Jinggouzi: archaeological report and comprehensive research on the late bronze age cemetery. Beijing: Science Press; 2010.
Institute of Cultural Relics and Archaeology of Inner Mongolia Ulanqab Museum. Administration of Cultural Relics of Huade County. The excavation of Xianbei cemeteries of Chengwugou site in Huade county, Inner Mongolian. Cao Yuan Wen Wu. 2014;1:45–48.
Liu G, Bai J, Chen Y, Li C, Shen R. The excavation of Gangga site in Chen Barag Banner, Inner Mongolian. Archaeology. 2015;7:75–86.
Kayser M, Brauer S, Cordaux R, Casto A, Lao O, Zhivotovsky LA, et al. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol Biol Evol. 2006;23:2234–44.
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–8.
Cui YQ, Li H, Ning C, Zhang Y, Chen L, Zhao X, et al. Y chromosome analysis of prehistoric human populations in the West Liao River Valley, Northeast China. BMC Evol Biol. 2013;13:216.
Wei LH, Huang YZ, Yan S, Wen SQ, Wang LX, Du PX, et al. Phylogeny of Y-chromosome haplogroup C3b-F1756, an important paternal lineage in Altaic-speaking populations. J Hum Genet. 2017;62:915–8.
Lkhagvasuren G, Shin H, Lee SE, Tumen D, Kim JH, Kim KY, et al. Molecular genealogy of a Mongol queen’s family and her possible kinship with Genghis Khan. PLoS ONE. 2016;11:e0161622. doi:10.1371/journal.pone.0161622
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31371266). We are grateful to Dr. Guoxiang Liu of the Institute of Archaeology, Chinese Academy of Social Sciences, and the Institute of Cultural and Historical Relics and Archaeology in the Inner Mongolia Nationality Autonomous Region in China for providing the samples.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
This study was approved by the Institute of Cultural and Historical Relics and Archaeology in the Inner Mongolia Nationality Autonomous Region in China, and the State Administration of Cultural Heritage of China. All necessary permits were obtained for the described study, which complied with all relevant regulations. We have no financial and personal relationships with people or organisations that can inappropriately influence our work.
Rights and permissions
About this article
Cite this article
Zhang, Y., Wu, X., Li, J. et al. The Y-chromosome haplogroup C3*-F3918, likely attributed to the Mongol Empire, can be traced to a 2500-year-old nomadic group. J Hum Genet 63, 231–238 (2018). https://doi.org/10.1038/s10038-017-0357-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-017-0357-z
This article is cited by
-
Ancient DNA from Tubo Kingdom-related tombs in northeastern Tibetan Plateau revealed their genetic affinity to both Tibeto-Burman and Altaic populations
Molecular Genetics and Genomics (2022)
-
Origin of ethnic groups, linguistic families, and civilizations in China viewed from the Y chromosome
Molecular Genetics and Genomics (2021)
-
Phylogenetic analysis of the Y-chromosome haplogroup C2b-F1067, a dominant paternal lineage in Eastern Eurasia
Journal of Human Genetics (2020)
-
Y-chromosomal analysis of clan structure of Kalmyks, the only European Mongol people, and their relationship to Oirat-Mongols of Inner Asia
European Journal of Human Genetics (2019)