Abstract
Current deep-sequencing technology provides a mass of nucleotide variations associated with human genetic disorders to accelerate the identification of causative mutations. To understand the etiology of genetic disorders, reverse genetics in human cultured cells is a useful approach for modeling a disease in vitro. However, gene targeting in human cultured cells is difficult because of their low activity of homologous recombination. Engineered endonucleases enable enhancement of the local activation of DNA repair pathways at the human genome target site to rewrite the desired sequence, thereby efficiently generating disease-modeling cultured cell clones. These edited cells can be used to explore the molecular functions of a causative gene product to uncover the etiological mechanisms. The correction of mutations in patient cells using genome editing technology could contribute to the development of unique gene therapies. This technology can also be applied to screening causative mutations. Rare genetic disorders and non-exonic mutation-caused diseases remain frontier in the field of human genetics as it is difficult to validate whether the extracted nucleotide variants are mutation or polymorphism. When isogenic human cultured cells with a candidate variant reproduce the pathogenic phenotypes, it is confirmed that the variant is a causative mutation.
Similar content being viewed by others
Introduction
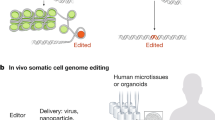
Both forward and reverse genetics approaches are required to understand the precise etiology of human genetic disorders. Forward genetics enables extraction of a causative gene mutation in patients, while reverse genetics allows the molecular functions of the causative gene product to be dissected to uncover the pathological mechanism. Via developments in deep-sequencing technology using next-generation sequencers, we have revealed and analyzed numerous nucleotide variations associated with human genetic diseases for efficient identification of the causative mutations [1, 2]. In contrast, reverse genetics in human cells for evaluating the molecular functions of these causative genes had been limited before genome editing technology was developed. As human cultured cells generally show low activity of homologous recombination, it was too difficult to generate disease-model cultured cells using the conventional method [3]. Engineered endonucleases (EENs) including ZFN, TALEN, and CRISPR/Cas9, increase the efficacy of genome editing through the site-specific activation of DNA repair activity to explore the reverse genetics in human cultured cells. Currently, it is practical for most researchers to dissect the molecular functions of causative gene products using edited cultured cell clones. As a therapeutic concept, if genome editing technology-mediated conversion of a mutation in patient cells to the reference sequence restores disease phenotypes, this has potential implications for the development of associated therapies (Fig. 1a). Further perspectives on genome editing technology mediated gene therapy have also been reviewed recently [4,5,6].
Strategies of genome editing technology in human cultured cells. a The available procedures in genome editing technology in medicine. Introduction of a gene mutation of interest into normal cultured cells is used to identify the molecular and cellular pathology for disease modeling. Another potential approach is to modify a mutation to the reference sequence in patient cells for gene therapy. b Error-prone NHEJ-mediated gene knockout. A target sequence is disrupted by an insertion or deletion due to NHEJ. c HDR-mediated introduction of a selectable drug resistance cassette (Drug R) into a target locus for gene knockout. d An NHEJ-mediated drug cassette with the same Cas9 recognition sequence as the endogenous target knock-in. When the Cas9 recognition sequence is in the same orientation in the endogenous target and drug cassette vector, the ends of the targeted vectors contain uncontrolled indels. In contrast, when the orientation of Cas9 recognition sequence in the endogenous target and drug cassette vector is opposed, the ends are programmable. e A simple SNV knock-in. A CRISPR/Cas9-induced DSB enhances HDR with a 100-nt ssODN repair template to introduce an SNV (asterisk) into a target site. f Site-specific cytidine deamination. Catalytically dead Cas9 protein recruits a cytidine deaminase AID (or APOBEC1) to specific sites, leading to conversion of C to T. g CORRECT method for SNV knock-in. In the first step, an SNP (asterisk) is introduced along with the second site mutations (closed circles) required to prevent recleavage by the Cas9 enzyme. A subsequent second step of editing in a similar manner corrects the secondary mutations to leave only the SNV. h Scarless SNV knock-in. A drug-selectable marker cassette is introduced into a target locus along with the SNV, and is subsequently excised by a further round of HDR or by piggyBack transposase
To obtain a precise understanding of the genetic basis of human diseases, it may be essential to use reverse genetics in human cultured cells. Genome editing technology enables engineering of the variant allele associated with a specific disease in human cultured cells with a uniform genetic background [7, 8]. Phenotypic comparison of such edited cells can then demonstrate how a variant of interest can affect the cellular events that are relevant to a specific pathological condition [9, 10]. If a knock-in variant reproduces disease-associated phenotypes, the causality of the candidate variant is confirmed (Fig. 1a). Here, we address the utility of employing genome editing technology in human cultured cells and discuss to what extent this technology can be applied in the field of human genetics.
Which human cultured cells are optimal for genome editing technology?
As a critical step of genome editing technology in human cultured cells, DNA double-stranded breaks (DSBs) induced by engineered endonucleases are repaired mainly by error-prone nonhomologous end joining (NHEJ) or error-free homologous directed repair (HDR) [11,12,13,14,15,16]. NHEJ, which is active throughout the cell cycle, causes insertions or deletions (indels) of various lengths that can lead to frameshift mutations and, consequently, gene knockout [17]. In contrast, HDR, which works in late-S and G2 phases, induces a precise recombination event between a homologous DNA donor and the DSB site, resulting in accurate introduction of the DNA donor into the target site and, consequently, gene knock-in [18]. Subsequently, the edited cells should be isolated and expanded for further functional evaluations. To distinguish clonal and artificial effects in the edited cell clones, it is necessary to generate a small number of independent edited clones, perform complementary analysis using exogenous expression of wild-type cDNA in the edited cells, or sequence the predicted off-target sites. Taking these points together, it is important to determine the DNA repair and proliferative capacities of the target cultured cells (Table 1).
Primary cells
A variety of human cultured cells have been used for the modeling of human diseases in vitro. Primary cells derived from patients, such as skin fibroblasts and peripheral lymphocytes, are informative to understand the cellular etiology of genetic disorders. However, the limited proliferative capacity of primary cells is a major obstacle in genome editing technology-mediated reverse genetics [19]. Thus, primary cells are currently not used for genome editing technology-mediated disease modeling. On the other hand, for the application of genome editing technology to gene therapy, primary cells are an essential target. Recently, Howden et al. [20] demonstrated the simultaneous reprogramming and genome editing of primary skin fibroblasts. However, further studies are needed to construct the experimental flow of genome editing technology in primary cells.
Cancer cell lines
Immortalized human cultured cell lines are useful for genome editing technology. Cancer cell lines are typically spontaneously immortalized cells [21,22,23]. As cancer cells have acquired unlimited proliferative ability via mutations in oncogenes and tumor suppressor genes during carcinogenesis, it is possible to generate immortalized cell lines from various cancer tissues [24]. However, most cancer cell lines show genomic instability, defined as either chromosomal instability (CIN) or microsatellite instability (MIN) [25]. CIN is characterized by aneuploidy due to chromosomal mis-segregation during mitosis [26, 27]. U2OS cells and HeLa cells, which are derived from osteosarcoma and cervical cancer, respectively, are typical examples of CIN [21, 28]. For genome editing, it is somewhat difficult to handle CIN cells because these cells have multiple copies of a target locus. In contrast, MIN is defined by repetitive DNA expansions without aneuploidy [29, 30]. A typical MIN cell line, HCT116, derived from colorectal carcinoma, has a stable karyotype of 45 chromosomes and relatively high activity of homologous recombination [31, 32]. Thus, HCT116 cells are often used for genome editing technology.
Immortalized normal cell lines
Cancer cell lines lose some morphological and biochemical characteristics observed in normal cells. For example, most cancer cell lines have lost extracellular signaling sensor structures, namely, primary cilia, which are hair-like, microtubule-based organelles present on the surface of most normal cells in the quiescent G0 phase [33]. To immortalize normal cells, exogenous expression of adenovirus type 5 E1 gene, simian virus 40 T-antigen, and/or human telomerase (hTERT) is used [34,35,36]. HEK293 cells, which are widely used in genome editing research, are adenovirus type 5 E1 gene-mediated immortalized human embryonic kidney cells [37]. Although the genomes of HEK293 cells can be readily edited because of the high rate with which transgenes can be introduced into them and their relaxed chromatin state, they lose their diploid karyotype and morphological features such as primary cilia and cell-to-cell junctions. In contrast, hTERT-RPE1 cells derived from normal human retina pigmented epithelia retain their original phenotypes of a diploid karyotype and primary cilium formation [38]. However, they also retain the low HDR activity as also observed in the original tissue [39]. For genome editing in hTERT-immortalized normal cells, we should choose an NHEJ-dependent editing strategy to improve the efficacy of isolation of the edited cell clones [40]. Taking these factors together, when establishing an experimental design, it is important to consider the DNA repair activity of the immortalized normal cells.
Induced pluripotent stem cells
Human pluripotent stem cells (hPSCs) are also important for genome editing technology, since they can continue to divide to form identical cell clones [7, 41, 42]. They can also produce specialized types of cells through differentiation. These properties of hPSCs confer a multitude of possibilities for the modeling of human diseases and the development of unique therapies. Human embryonic stem cells (hESCs), derived from the inner cell mass of the blastocyst during human embryogenesis, were originally referred to as hPSCs [43]. However, the generation of hESCs is inherently associated with challenges in accessing fertilized eggs and ethical issues regarding their use.
To overcome these issues, in 2006, iPS technology emerged to provide a robust approach for generating hPSCs without the use of embryos [44]. In iPS technology, adult somatic cells transfected by the four Yamanaka factors, including Oct3/4, Sox2, c-Myc, and Klf4, can be reprogrammed to acquire stem cell characteristics [44, 45]. Induced pluripotent stem cells (iPSCs) generated from normal individuals have been verified to have a diploid karyotype, hESC-like DNA methylation pattern, and potential to develop into all three germ layers [45]. An intrinsic feature of single-cell survival rate in iPSCs is a challenge for reverse genetics. The suppression of anoikis by the Rho-kinase inhibitor Y-27632 during the disaggregation of hESC colonies was found to dramatically improve single-cell survival of hESCs, so it can be applied to single-cell cloning of iPSCs [46, 47]. Genome editing technology has been combined with iPSCs to generate knockout or knock-in clones, correct causative mutations, or insert reporter genes [48,49,50,51].
Which genome editing method is used for human genetics studies?
There are numerous strategies for genome editing in human cultured cells. The choice of such a strategy depends on the specific issue that is being addressed (knockout or knock-in) and multiple experimental factors including the types of human cultured cells, EENs, and transfection methods [52]. Therefore, it is necessary to optimize the experimental flow in view of the specific aim. Here, we summarize successful examples of genome editing technology-mediated strategies applied to human genetics studies (Fig. 1b–h, Table 2).
Gene knockout in human cultured cells
Gene knockout is a simple but important approach for modeling a disease. EEN-mediated NHEJ introduces indel mutations into a target locus (Fig. 1b). These indel mutations in protein-coding regions cause frameshifts to generate a null-mutant cell. To date, many disease-model cells generated by this strategy have been reported [53,54,55,56,57]. In the field of human genetics, these null-mutant cells are also used in complementary tests for the candidate variants underlying a genetic disorder called by forward genetics approach.
Using a homology arm-tagged targeting vector and EENs, HDR leads to the replacement of a protein-coding exon with a drug resistance cassette for the efficient isolation of a knockout cell (Fig. 1c) [39, 58, 59]. When targeting arms containing flanking constitutive exons with recombinase sites such as loxP sites are used in this strategy, cells with conditional knockout in the genes essential for cell survival can be generated [60, 61]. Moreover, EEN-mediated incorporation of a targeting vector possessing the desired variant in the homology arms into the target locus enables modeling or correction of disease-associated phenotypes in vitro [62, 63].
NHEJ-mediated knock-in using CRISPR-ObLiGaRe or HITI
Genome editing technology has mainly been applied to human cell lines with intrinsic HDR activity that is sufficient for the efficient isolation of genome-edited cells. However, HDR-dependent genome editing is not practical in normal-tissue-derived cell lines and post-mitotic cells including neurons and muscle cells since their HDR activity is inefficient or deficient [64, 65]. For example, we previously generated a microcephaly-associated KIF2A gene knockout hTERT-RPE1 cell line using TALEN and a drug-resistant gene cassette contained in a targeting vector, but the efficacy of isolation of the targeted clones was low, at approximately 1% of drug-resistant clones [39]. Maresca et al. added the ZFN site located in the genome into a drug-resistant gene cassette vector, and cointroduced the ZFN and the targeting vector into human cultured cells to isolate the targeted clones with high efficacy through NHEJ activity (Fig. 1d) [66]. They named this method ObLiGaRe (obligate ligation-gated recombination), based on the Latin verb obligate (“to join to”) [66]. In the ObLiGaRe method, highly effective transgene knock-in occurs in most human cultured cells, but the orientation of the transgene is not controlled and precise adjustment of junction sequences between a target locus and the transgene is not possible. Recently, we combined the CRISPR/Cas9 system and the ObLiGaRe method to efficiently generate ataxia-telangiectasia-causative ATM gene knockout hTERT-RPE1 cell clones [67]. In this method, biallelic targeting vector-inserted clones corresponding to knockout cells were rare at around 5% among the drug-resistant clones, while the monoallelic inserted clones were dominant at more than 70%. As almost all monoallelic inserted clones carried the NHEJ-mediated insertions or deletions at the target locus in the second, uninserted allele, >70% of the drug-resistant clones were indeed knockout cell clones. Thus, CRISPR-ObLiGaRe is an efficient and useful method for generating knockout cell clones.
Notably, a novel NHEJ-mediated site-specific transgene insertion method named homology-independent targeted integration (HITI) has been reported [68]. In CRISPR-ObLiGaRe, a guide RNA (gRNA) target sequence located in the genome is added in the same orientation into the targeting vector [66, 67]. In contrast, the targeting vector for HITI contains the gRNA target sequence in the opposite orientation to the genome [68]. HITI-mediated transgene knock-in occurs more preferentially in the forward than in the reverse direction because the forward-directed transgene knock-in alters the genomic gRNA target sequence to prevent additional CRISPR/Cas9 cutting. Suzuki et al. demonstrated that HITI worked in HEK293 cells and post-mitotic neurons, and that HITI introduced the wild-type exon to rescue visual function using a rat model of retinitis pigmentosa as a proof of concept of its potential use for gene correction therapy [68].
ssODN-mediated single nucleotide variation (SNV) knock-in
Numerous SNVs have been identified from the screening of causative mutations in genetic disorders and Genome-wide association study (GWAS) [1, 2, 69, 70]. To validate the causality of these SNVs, EEN-driven HDR introduces 100–200 nt of single-stranded DNA oligonucleotides (ssODN) with the SNV into the target locus (Fig. 1e) [48, 71]. Generally, the distance of SNV from the DSB should be minimized for efficient SNV incorporation [72, 73]. This method is routinely performed in early embryos of many animal species [74,75,76]. Although this method was reported to have been applied to mutation correction in some patient-derived iPSCs [50, 77, 78], it is not efficient in human cultured cells because of the less efficient HDR activity. Therefore, it is necessary to design experimental procedures for the concentration of ssODN-knock-in cells. To date, sib-selection and transient drug selection methods to achieve this concentration have been reported [52, 79], but further improvements are needed for practical use.
Site-specific cytidine deamination as a scarless SNV knock-in
The missense mutations of both D10A and H840A in the Cas9 protein inactivate its nuclease activity, while retaining its ability to bind to specific DNA sequences [80]. Conjugation of such catalytically dead Cas9 (dCas9) with an alternative enzymatic domain allows the recruitment of specific enzymatic activities to the target site in the genome [81,82,83,84,85,86]. This has been applied for the conversion of one base to another directly in the target genome. Activation-induced cytidine deaminase (AID) and dCas9 were fused to form a synthetic complex named Target-AID that converts C to T (or G to A) at the specific base (Fig. 1f) [87]. Another cytidine deaminase, APOBEC1, is also available for the programmable base editing mediated by dCas9 technology (Fig. 1f) [88]. In addition, in a cancer cell line, HCC1954, with Tyr163Cys mutation in the tumor suppressor gene p53, dCas9-APOBEC1 corrected the mutation by a specific C-to-T transition at a rate of 3.3%–7.6% [89]. Since this approach does not require any DNA repair activity, it can be applied to a broad range of cell types. However, it requires a procedure for concentrating single-base-pair-substituted cells similarly to the case of ssODN-knock-in cells. In addition, since only C-to-T transition is currently available in this approach, its application is somewhat limited.
CORRECT method for scarless SNV knock-in
A two-step genome editing strategy named “CORRECT” for scarless SNV knock-in has been reported (Fig. 1g) [90, 91]. In the first step of this approach, to prevent recutting by CRISPR/Cas9 and unwanted indels subsequently being introduced into human cultured cells using an ssODN template carrying the intended mutation and secondary silent mutations. In the second step, only the secondary mutations are removed by a redesigned guide RNA, which targets the 20-bp sequence containing the introduced CRISPR/Cas9-blocking mutations and the modified repair ssODN template. Alternatively, a Cas9 variant such as VRER-Cas9, which recognizes the modified PAM sequence introduced as a blocking mutation in the first step, can also be used in the second step [90].
Using this approach, early-onset Alzheimer’s disease-causative mutations in amyloid precursor protein (APPSwe) and presenilin1 (PSEN1M146V) were precisely introduced into HEK293 cells and iPSCs [90]. The edited iPSC-derived cortical neurons displayed the disease-specific biochemical phenotypes of amyloid-β (A-β) peptide generation [90]. Thus, Alzheimer’s disease-associated phenotypes in neurons can be faithfully modeled in vitro using genome editing technology in human iPSCs. However, this approach requires at least two rounds of clonal selection, taking ~3 months to generate the intended mutant in iPSCs. To minimize clonal selection and the occurrence of indels at each step, HDR improvement strategies such as novel NHEJ inhibitors and HDR enhancers should be applied in this approach.
Drug-selectable scarless SNV knock-in
For efficient SNV introduction in human cultured cells, we previously reported a TALEN-mediated two-step single-base-pair editing strategy (Fig. 1h) [92]. The first step included TALEN-mediated insertion of a drug-selectable marker cassette into an SNV flanking region. The targeting vector carried a neomycin-resistance gene and a herpes simplex virus thymidine kinase (hsvTK) gene separated by a 2A peptide sequence, allowing expression of the discrete protein products from a single open reading frame. The drug-selectable marker cassette knock-in cell clones were positively selected using neomycin. The second step involved the removal of the drug-selectable marker cassette from the targeted alleles, and introduction of the single-nucleotide substitution in an HDR-activity-dependent manner. Single-nucleotide-edited clones were negatively selected using ganciclovir treatment. Compared with the CORRECT method, the TALEN-mediated two-step single-base-pair editing strategy enables more efficient isolation of the edited clones, since it uses an antibiotic resistance marker for screening. Using this approach, we identified a causal mutation of a cancer-prone genetic disorder, premature chromatid separation with mosaic variegated aneuploidy [PCS (MVA)] syndrome [92,93,94]. Both biallelic and monoallelic mutations of the BUB1B gene encoding BubR1 have been reported in patients [95, 96]. Monoallelic mutations in the exons of BUB1B were identified in seven Japanese families with this syndrome. No second mutation in exons of the BUB1B gene was found in the opposite allele, although a conserved BUB1B haplotype within a 200-kb interval among the Japanese patients and a reduced transcript level were identified [96]. We thus determined the nucleotide sequence of the 200-kb region in a patient with this syndrome using deep-sequencing analysis and found that a unique SNV in an intergenic region 44-kb upstream of the BUB1B transcription start site was linked to the disease [92]. We used TALEN-mediated single-base-pair editing technology to biallelically introduce this substitution into HCT116 cells. The genome-edited clones showed reduced BUB1B transcript levels and PCS (MVA) syndrome-specific chromosomal instability, demonstrating that the identified SNV was indeed the causal mutation [92]. The single-base-pair editing technique is thus useful to evaluate nucleotide variants with unknown functional relevance.
In this approach, TALEN can be replaced by other EENs including CRISPR/Cas9. Moreover, the drug-selectable cassette marker can be excised either by site-specific recombinases or by the piggyBac transposase (Fig. 1h), which has recently been demonstrated to be effective in human iPSCs [51, 97,98,99,100]. However, the targetable loci are limited since editing with piggyBac transposase requires a TTAA sequence near the target site [98]. These strategies are useful for efficient SNV knock-in in human cultured cells, but they require at least two rounds of clone selection, which is time-consuming and can produce off-target mutations. Further investigations are thus needed to achieve safe scarless SNV-knock-in systems.
Conclusions
Genome editing technology considering each cellular characteristic will undoubtedly provide major insights into the pathological mechanisms and therapeutic targets of genetic disorders. However, several important challenges remain. The most critical limitation in this technology is derived from the general property of human cultured cells in which their DSBs are repaired dominantly by the NHEJ pathway rather than by HDR. Therefore, an NHEJ-mediated gene knockout strategy is mainly used to evaluate the loss-of-function effects and to remove the disease-causing sequences. In the case of Duchenne muscular dystrophy (DMD) patient-derived iPSCs, a 725-kb genomic region containing a premature stop codon in the disease-causing DMD gene was deleted by CRISPR/Cas9 to rescue the open reading frame and ensure partial protein function [101]. In addition, to treat HIV-infected patients, the CCR5 gene, which encodes a chemokine co-receptor required for HIV infection but not survival, of T cells was removed by ZFN, for which clinical trials are underway [102]. However, for most clinical treatments or precise modeling of diseases in vitro, it is essential to achieve high-frequency knock-in of the repair template with the desired variant. Notably, since microhomology-mediated end joining (MMEJ)-assisted gene knock-in named PITCh (Precise Integration into Target Chromosome) [103, 104] and NHEJ-mediated site-specific gene insertion, HITI [68], are both HDR-independent precise knock-in methods, these strategies may increase the utility of genome editing in human cultured cells. Other techniques to enhance gene knock-in include inhibition of NHEJ with small compounds or Cas9 protein accumulation in an S- and G2-phase-dependent manner [105,106,107,108,109]. Several techniques to shift the DSB repair balance from NHEJ to HDR in human cultured cells have been developed. A class2 CRISPR effector, Cpf1, which cuts target DNA further away from the PAM sequence to generate a single-stranded overhang, may increase HDR more than NHEJ [110]. Against this background, further advances of genome editing technology in human cultured cells are required to understand and correct genetic disorders.
References
Deciphering Developmental Disorders, S. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–8.
Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–14.
Sedivy JM, Vogelstein B, Liber HL, Hendrickson EA, Rosmarin AG. Gene targeting in human cells without isogenic DNA. Science. 1999;283:9.
Cornu TI, Mussolino C, Cathomen T. Refining strategies to translate genome editing to the clinic. Nat Med. 2017;23:415–23.
Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121–31.
Maeder ML, Gersbach CA. Genome-editing technologies for gene and cell therapy. Mol Ther. 2016;24:430–46.
Bassett, A.R. Editing the genome of hiPSC with CRISPR/Cas9: disease models. Mamm Genome. 2017;28:348–364.
Yang L, Yang JL, Byrne S, Pan J, Church GM. CRISPR/Cas9-directed genome editing of cultured cells. Curr Protoc Mol Biol. 2014;107:31.
Urnov FD. Human genome editing as a tool to establish causality. Proc Natl Acad Sci USA. 2014;111:1233–4.
Merkle FT, Eggan K. Modeling human disease with pluripotent stem cells: from genome association to function. Cell Stem Cell. 2013;12:656–68.
Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–46.
Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55.
Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet. 2014;23:R40–6.
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23.
Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18:495–506.
Renkawitz J, Lademann CA, Jentsch S. Mechanisms and principles of homology search during recombination. Nat Rev Mol Cell Biol. 2014;15:369–83.
Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621.
Howden SE, Maufort JP, Duffin BM, Elefanty AG, Stanley EG, Thomson JA. Simultaneous reprogramming and gene correction of patient fibroblasts. Stem Cell Rep. 2015;5:1109–18.
Scherer WF, Syverton JT, Gey GO. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med. 1953;97:695–710.
Shay JW, Tomlinson G, Piatyszek MA, Gollahon LS. Spontaneous in vitro immortalization of breast epithelial cells from a patient with Li-Fraumeni syndrome. Mol Cell Biol. 1995;15:425–32.
Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–74.
Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67.
Draviam VM, Xie S, Sorger PK. Chromosome segregation and genomic stability. Curr Opin Genet Dev. 2004;14:120–5.
Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, et al. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–3.
Funk LC, Zasadil LM, Weaver BA. Living in CIN: mitotic infidelity and its consequences for tumor promotion and suppression. Dev Cell. 2016;39:638–52.
Ponten J, Saksela E. Two established in vitro cell lines from human mesenchymal tumours. Int J Cancer. 1967;2:434–47.
Strand M, Prolla TA, Liskay RM, Petes TD. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–6.
Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70.
Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–501.
Roschke AV, Stover K, Tonon G, Schaffer AA, Kirsch IR. Stable karyotypes in epithelial cancer cell lines despite high rates of ongoing structural and numerical chromosomal instability. Neoplasia. 2002;4:19–31.
Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–78.
Speiseder, T., Hofmann-Sieber, H., Rodriguez, E., Schellenberg, A., Akyuz, N., Dierlamm, J. et al. Efficient transformation of primary human mesenchymal stromal cells by adenovirus early region 1 oncogenes. J Virol. 2017:91:e01782–16.
Matsumura T, Takesue M, Westerman KA, Okitsu T, Sakaguchi M, Fukazawa T, et al. Establishment of an immortalized human-liver endothelial cell line with SV40T and hTERT. Transplantation. 2004;77:1357–65.
Lundberg AS, Hahn WG, Gupta P, Weinberg RA. Genes involved in senescence and immortalization. Curr Opin Cell Biol. 2000;12:705–9.
Dubridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. Analysis of mutation in human-cells by using an Epstein-Barr-Virus shuttle system. Mol Cell Biol. 1987;7:379–87.
Jiang XR, Jimenez G, Chang E, Frolkis M, Kusler B, Sage M, et al. Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat Genet. 1999;21:111–4.
Miyamoto T, Hosoba K, Ochiai H, Royba E, Izumi H, Sakuma T, et al The microtubule-depolymerizing activity of a mitotic kinesin protein KIF2A drives primary cilia disassembly coupled with cell proliferation. Cell Rep. 2015;S2211-1247:00004–2.
Katoh Y, Michisaka S, Nozaki S, Funabashi T, Hirano T, Takei R, et al. Practical method for targeted disruption of cilia-related genes by using CRISPR/Cas9-mediated, homology-independent knock-in system. Mol Biol Cell. 2017;28:898–906.
Muffat J, Li Y, Jaenisch R. CNS disease models with human pluripotent stem cells in the CRISPR age. Curr Opin Cell Biol. 2016;43:96–103.
Parekh U, Yusupova M, Mali P. Genome engineering in human pluripotent stem cells. Curr Opin Chem Eng. 2017;15:56–67.
Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7.
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76.
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72.
Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–6.
Ohgushi M, Matsumura M, Eiraku M, Murakami K, Aramaki T, Nishiyama A, et al. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell. 2010;7:225–39.
Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–7.
Liu GH, Suzuki K, Qu J, Sancho-Martinez I, Yi F, Li M, et al. Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell Stem Cell. 2011;8:688–94.
Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–31.
Wang G, Yang L, Grishin D, Rios X, Ye LY, Hu Y, et al. Efficient, footprint-free human iPSC genome editing by consolidation of Cas9/CRISPR and piggyBac technologies. Nat Protoc. 2017;12:88–103.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.
Yost S, de Wolf B, Hanks S, Zachariou A, Marcozzi C, Clarke M, et al. Biallelic TRIP13 mutations predispose to Wilms tumor and chromosome missegregation. Nat Genet. 2017;49:1148–51.
Liu Z, Hui Y, Shi L, Chen Z, Xu X, Chi L, et al. Efficient CRISPR/Cas9-mediated versatile, predictable, and donor-free gene knockout in human pluripotent stem cells. Stem Cell Rep. 2016;7:496–507.
Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616–23.
Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet. 2015;47:469–78.
Horii T, Tamura D, Morita S, Kimura M, Hatada I. Generation of an ICF syndrome model by efficient genome editing of human induced pluripotent stem cells using the CRISPR system. Int J Mol Sci. 2013;14:19774–81.
Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–8.
Li Y, Wang H, Muffat J, Cheng AW, Orlando DA, Loven J, et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13:446–58.
Economides AN, Frendewey D, Yang P, Dominguez MG, Dore AT, Lobov IB, et al. Conditionals by inversion provide a universal method for the generation of conditional alleles. Proc Natl Acad Sci USA. 2013;110:E3179–88.
Andersson-Rolf A, Mustata RC, Merenda A, Kim J, Perera S, Grego T, et al. One-step generation of conditional and reversible gene knockouts. Nat Methods. 2017;14:287–9.
Li H, Bielas SL, Zaki MS, Ismail S, Farfara D, Um K, et al. Biallelic mutations in citron kinase link mitotic cytokinesis to human primary microcephaly. Am J Hum Genet. 2016;99:501–10.
Chang CW, Lai YS, Westin E, Khodadadi-Jamayran A, Pawlik KM, Lamb LS Jr., et al. Modeling human severe combined immunodeficiency and correction by CRISPR/Cas9-enhanced gene targeting. Cell Rep. 2015;12:1668–77.
Pardo B, Gomez-Gonzalez B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci. 2009;66:1039–56.
Heidenreich M, Zhang F. Applications of CRISPR-Cas systems in neuroscience. Nat Rev Neurosci. 2016;17:36–44.
Maresca M, Lin VG, Guo N, Yang Y. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013;23:539–46.
Royba E, Miyamoto T, Akutsu S, Hosoba K, Tauchi H, Kudo Y, et al. Evaluation of ATM heterozygous mutations underlying individual differences in radiosensitivity using genome editing in human cultured cells. Sci Rep. 2017;7:5996.
Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–9.
Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet 2012;13:135–45.
Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, et al. 10 Years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101:5–22.
Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8:753–5.
Li HL, Gee P, Ishida K, Hotta A. Efficient genomic correction methods in human iPS cells using CRISPR-Cas9 system. Methods. 2016;101:27–35.
Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–61.
Yoshimi K, Kunihiro Y, Kaneko T, Nagahora H, Voigt B, Mashimo T. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun. 2016;7:10431.
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8.
Armstrong GA, Liao M, You Z, Lissouba A, Chen BE, Drapeau P. Homology directed knockin of point mutations in the zebrafish tardbp and fus genes in ALS Using the CRISPR/Cas9 system. PLoS ONE. 2016;11:e0150188.
Li C, Ding L, Sun CW, Wu LC, Zhou D, Pawlik KM, et al. Novel HDAd/EBV reprogramming vector and highly efficient Ad/CRISPR-Cas sickle cell disease gene correction. Sci Rep. 2016;6:30422.
Niu X, He W, Song B, Ou Z, Fan D, Chen Y, et al. Combining single strand oligodeoxynucleotides and CRISPR/Cas9 to correct gene mutations in beta-thalassemia-induced pluripotent stem cells. J Biol Chem. 2016;291:16576–85.
Miyaoka Y, Chan AH, Judge LM, Yoo J, Huang M, Nguyen TD, et al. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat Methods. 2014;11:291–3.
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–83.
Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol. 2016;17:5–15.
Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–51.
Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–91.
Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci USA. 2015;112:3002–7.
Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, et al. Editing DNA methylation in the mammalian genome. Cell. 2016;167:233–47 e217.
Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, et al. Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat Biotechnol. 2016;34:1060–5.
Nishida, K., Arazoe, T., Yachie, N., Banno, S., Kakimoto, M., Tabata, M. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016:353:aaf8729.
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–4.
Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods. 2016;13:1029–35.
Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–9.
Kwart D, Paquet D, Teo S, Tessier-Lavigne M. Precise and efficient scarless genome editing in stem cells using CORRECT. Nat Protoc. 2017;12:329–54.
Ochiai H, Miyamoto T, Kanai A, Hosoba K, Sakuma T, Kudo Y, et al. TALEN-mediated single-base-pair editing identification of an intergenic mutation upstream of BUB1B as causative of PCS (MVA) syndrome. Proc Natl Acad Sci USA. 2014;111:1461–6.
Kajii T, Kawai T, Takumi T, Misu H, Mabuchi O, Takahashi Y, et al. Mosaic variegated aneuploidy with multiple congenital abnormalities: homozygosity for total premature chromatid separation trait. Am J Med Genet. 1998;78:245–9.
Matsuura S, Ito E, Tauchi H, Komatsu K, Ikeuchi T, Kajii T. Chromosomal instability syndrome of total premature chromatid separation with mosaic variegated aneuploidy is defective in mitotic-spindle checkpoint. Am J Hum Genet. 2000;67:483–6.
Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–61.
Matsuura S, Matsumoto Y, Morishima K, Izumi H, Matsumoto H, Ito E, et al. Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. Am J Med Genet A. 2006;140:358–67.
Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–4.
Yusa K. Seamless genome editing in human pluripotent stem cells using custom endonuclease-based gene targeting and the piggyBac transposon. Nat Protoc. 2013;8:2061–78.
Imamura K, Sahara N, Kanaan NM, Tsukita K, Kondo T, Kutoku Y, et al. Calcium dysregulation contributes to neurodegeneration in FTLD patient iPSC-derived neurons. Sci Rep. 2016;6:34904.
Maetzel D, Sarkar S, Wang H, Abi-Mosleh L, Xu P, Cheng AW, et al. Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells. Stem Cell Rep. 2014;2:866–80.
Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, et al. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell. 2016;18:533–40.
Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901–10.
Nakade S, Tsubota T, Sakane Y, Kume S, Sakamoto N, Obara M, et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun. 2014;5:5560.
Sakuma T, Nakade S, Sakane Y, Suzuki KT, Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc. 2016;11:118–33.
Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–42.
Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–8.
Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–7.
Song J, Yang D, Xu J, Zhu T, Chen YE, Zhang J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun. 2016;7:10548.
Gutschner T, Haemmerle M, Genovese G, Draetta GF, Chin L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep. 2016;14:1555–66.
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–71.
Acknowledgements
This work was supported by AMED-PRIME from the Japan Agency for Medical Research and Development, AMED (to TM); Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to TM and SM); Grant-in-Aid for Scientific Research from the Ministry of Health, Labor and Welfare (to SM); the Center of World Intelligence Projects for Nuclear S&T and Human Resource Development from the Japan Science and Technology Agency (to S.M.); research grants from the Naito Foundation (to SM and TM); the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to TM); Ono Medical Research Foundation (to TM); and Takeda Science Foundation (to TM).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Miyamoto, T., Akutsu, S. & Matsuura, S. Updated summary of genome editing technology in human cultured cells linked to human genetics studies. J Hum Genet 63, 133–143 (2018). https://doi.org/10.1038/s10038-017-0349-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-017-0349-z
This article is cited by
-
Disruptive Technology: CRISPR/Cas-Based Tools and Approaches
Molecular Diagnosis & Therapy (2019)