Abstract
Understanding the basis of regeneration of each tissue and organ, and incorporating this knowledge into clinical treatments for degenerative tissues and organs in patients, are major goals for researchers in regenerative biology. Here we provide an overview of current work, from high-regeneration animal models, to stem cell-based culture models, transplantation technologies, large-animal chimeric models, and programmable nuclease-based genome-editing technologies. Three-dimensional culture generating organoids, which represents intact tissue/organ identity including cell fate and morphology are getting more general approaches in the fields by taking advantage of embryonic stem cells, induced pluripotent stem cells and adult stem cells. The organoid culture system potentially has profound impact on the field of regenerative medicine. We also emphasize that the large animal model, in particular pig model would be a hope to manufacture humanized organs in in vivo empty (vacant) niche, which now potentially allows not only appropriate cell fate identity but nearly the same property as human organs in size. Therefore, integrative and collaborative researches across different fields might be critical to the aims needed in clinical trial.
Similar content being viewed by others
Introduction
Tissues and organs in the body perform essential bioactivity over the life. A sensory nervous system such as brain and retina play roles in recognition and visual functions. A digestive system, which consists of the gastrointestinal tract plus the accessory organs of digestion (tongue, salivary glands, pancreas, liver, and gallbladder) is critical to maintain a homeostasis, including a nutrient uptake. A circulation system such as heart, kidneys, and vasculature is also important for a fluid circulation, including cells, nutrients, gases and waste products. Loss of those systems by accidents and diseases could lead to life-threatening (or lethal) problems and loss of life quality.
The self-renewal process including self-repair is known to be essential for tissue turn over and tissue restoration upon degeneration and injury. For the replacement or self-restoration of damaged tissues important for homeostasis, a better understanding of the mechanism of regeneration is important. Over the last few decades, researchers have investigated model species that have highly regenerative tissues. To this end, we realize that the ability to return to the embryonic state, alternatively a regenerative ability in mammalian species is not as advanced as it is in fishes, amphibians, and birds although the morphology, the cell types, and the function of the several organs are relatively conserved among vertebrates. Therefore, to improve clinical treatments in regenerative medicine, we need to learn not only about highly regenerative mechanisms but also about those missing in less regenerative mammalian species, such as humans. Generally speaking, in humans, the loss of a body part requires external sources to partially replace and recover the original function. One such approach is transplantation-mediated replacement with full (or parts of) organs from a donor. Since donor organs are in short supply, alternative approaches that allow us to produce tissues and organs are needed.
Recent progress in the biotechnologies is providing new approaches to regenerative medicine that mainly fall into five categories: formation of tissues and organs from stem cells or tissues in a dish [1,2,3], stem-cell derived humanized organs in large animal models, three-dimensional (3D) bioprinting using biocompatible materials [4], ex vivo decellularization [5,6,7], and Xenotransplantation [8,9,10]. We do not discuss 3D bioprinting and decellularization technologies in detail in this review.
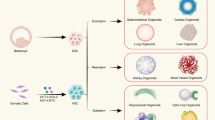
The use of embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) as the source of customized organs has become much more achievable than it was decades ago. For example, starting from a floating culture of ESCs (or iPSCs) in serum-containing media, called embryoid body culture, ESC/iPSCs can differentiate into specific cell types with individual marker expression characteristic of the three germ layers in vivo (Fig. 1). This differentiation ability, or pluripotency, is fundamental and essential for generation of desired tissues and organs.
Stem cell resources and experimental models using in vitro culture. a In normal development, a zygote becomes a blastocyst, which contains an inner cell mass (ICM), a body proper. Through a gastrulation, the ICM generates three germ layers: ectoderm, mesoderm, and endoderm. These layers further develop (or differentiate) into each organ until adulthood. Embryonic stem cells (ESCs) are derived from the ICM in the blastocyst. Induced pluripotent stem cells (iPSCs) can be generated from adult cells, such as dermal, blood, and photoreceptor cells, using Yamanaka factors (reprogramming factors) through a reprogramming process. b These pluripotent stem cells can be maintained in culture conditions and have an ability to differentiate into the three germ layers in response to specific stimulation such as growth factors. Adult stem cells (ASCs) have a restricted ability to generate specific tissues or organs in vitro with their original identity. c As an example, pluripotent stem cell (PSC)-derived eye organoids have been used in several experimental models, such as eye specification, morphogenesis, dorsal (D ventral (V) patterning, transdifferentiation, and disease models. NR, neuroretina; RPE, retinal pigment epithelium
Adult stem cells (ASCs), stem cells with a specific lineage, also have a remarkable ability to generate tissues with specific architecture when given an appropriate in vitro environment (Fig. 1). Substantial progress in stem cell culture techniques have been achieved recently, due to our better understanding of development in vivo: cell-type specification, embryonic cell-specific roles and crucial genes have been discovered by gene loss- and gain-of-function approaches. Thus, in vitro approaches using primary cells (tissues/organs) and cell lines have seen advances in how we manipulate and culture specific cells, tissues and organs using defined conditions to maintain their fate (or lineage) during growth.
There have been some remarkable developments in the use of human patient-derived cells as well. These stem cells are now considered to be a potentially valuable resource for regenerative approaches, as the problems of immune rejection and quantity of supply would be avoided. However, some roadblocks still remain, such as the fact that patient-derived cells are likely to have intrinsic genetic disorders that should be rescued or modified before their treatment.
Programmable nuclease-based genome editors, or so-called genome editing, are now recognized as a realistic genetic tool for curing diseases that are difficult to treat using currently available technologies. Genome editors are developing rapidly and in a diverse array of contexts. Zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR associated proteins 9 (CRISPR/Cas9) are well-known tools [11]. They can create random insertion or deletion of DNA, precise base pair changes, large deletions, inversions or duplications. Alternatively, a knock-in of a therapeutic transgene at a desired location, chromosomal rearrangement, epigenetic modifications, and gene activation (or repression) in transcriptional level is also possible [12,13,14,15,16,17,18,19,20] (Fig. 2a).
In vitro and in vivo organ generation, and genome editing. a There are three genome-editing tools, ZFNs, TALENs, and CRISPR/Cas9. Taking advantage of genome-editing technologies in several different ways will allow us to perform better experiments, and lead to better ideas for regenerative medicine. Validation of the roles of gene function: gain of function and loss of function. Further modulation of additional genes potentially rescue an abnormal phenotype: functional repair in genome level. By modulating essential factors found in vivo studies in a time- and dose-dependent manner, stem cell-derived tissues and organs can be now generated even in vitro. Genome-edited pluripotent stem cells (PSCs) are also useful for monitoring a specific cell lineage tracing, disease models, and developmental analysis, which might be one of the standard ways for understanding pathogenesis. The resultant knowledge obtained in vitro would help to perform better treatments in future clinical setting. b A in vivo humanized organ model, a large animal pig lacking (or modifying) an essential gene for pancreas development provide an empty (vacant) niche for human pluripotent stem cell-derived pancreas production in vivo. The genome-editing technology would be combined with the organ production technology outside a human body. Then, a better transplantation method for each organ would be necessary once individual humanized organs are mature enough to implant into patients. hiPSC, human iPSC. c Human-derived somatic cells can be used for iPSCs generation using reprogramming methods. iPSCs and adult stem cells (ASCs) from patients would have a cause of disease, which now can be repaired by genome-editing tools in clinical use. Even drug treatment might lessen the severity of diseases. Recent efforts are underway to develop in vivo genome editing, for more direct and local repair in the human body. ESCs, embryonic stem cells; RPE, retinal pigment epithelium
Among them, CRISPR/Cas9 is currently the most popular tool [21, 22], and the winner of Science Magazine’s Breakthrough of the 2015 Year award [23]. Since CRISPR loci were first described in 1987 [24], this system has been discovered to be necessary for acquired resistance against phages in bacteria and archeae [25]. It has turned out to be a bacterial immune system, which cleaves an invading foreign DNA in a sequence-specific manner [26]. Several experimental applications have been described [27] and they would be beneficial for biology, biotechnology, and biomedicine [28]. Further potential applications of the CRISPR/Cas9 tool include human gene therapy, viral gene disruption, agriculture, and next-generation therapies [28].
Arbitrarily introduced mutations using genome-editing tools alone showed recapitulation of several developmental abnormalities and diseases, including cancer. In contrast, by using a single-stranded oligonucleotide or double-stranded oligonucleotide as a repair template, these genome-editing tools have great potential for repairing patient-derived genetic mutations. To date, genome editing has worked in most well-studied and genome-deciphered model organisms and mammalian culture cells, including human cells [29,30,31,32,33,34,35,36,37,38,39,40]. The protocol for each of these model organisms is now being standardized. The combination of genome-editing and stem cell technologies might be quite essential for regenerative medicine.
In this review, we first describe regeneration (using the retina as an example). Then we focus on several in vitro methods for organ generation, and finally we discuss in vivo organ generation in large animal models and the use of genome-editing technologies as potential approaches toward regeneration and regenerative medicine.
Tissue and organ regeneration
Tissue turn over normally requires tissues or cells equivalent to bipotent or multipotent stem cells in order to provide newly generated cells. Regeneration of a complex system where acquisition of stemness from quiescent cells through a dedifferentiation (or reprogramming) process can be seen in the retina [41,42,43,44] and heart [45, 46]. The process requires reversion of a differentiated cell to a less differentiated state to allow proliferation or differentiation.
Mammalian species have limited regenerative capacity while some non-mammalian species like fishes and amphibians are known to have remarkable regenerative abilities. In highly regenerative model animals, heart and retina are used to identify genes and fundamental mechanisms for basic understanding of the regeneration; perhaps those mechanisms are useful for mammalian regeneration [47, 48].
Although the liver in mammals has a regeneration ability that is equivalent to other species after injury [49], the heart in mice has a restricted ability to regenerate after damage by neonatal day 7 [50, 51]. Recently Kenneth D. Poss and colleagues provided new insight in heart tissue regeneration, identifying tissue regeneration enhancer elements (TREEs) upon injury suggesting TREEs would be useful to modulate the regenerative potential of vertebrate organs [52]. Introduction of genes important for regeneration, such as Nrg1, using enhancer-based targeting by genome editing, might trigger repair processes even in the mammalian heart [52, 53].
The retina is an important sensory organ in vision. Vision loss by retinal degeneration is a critical issue that reduces the quality of life. There are three resources for the potential regeneration of retina (reviewed in [54–56]). First, retinal pigment epithelium (RPE) located outside the retina could transdifferentiate into the retina only in the amphibian upon removal of retina [57,58,59]; transdifferentiation is including a regenerative phenomenon where one cell type converts to another [60]. Second, ciliary marginal zone (CMZ) functions located at the rim of the retina are known to provide retinal neurons to the peripheral retina in larval and adult fish [61, 62]. Interestingly, several reports imply the stem-like cells in the mammalian CMZ as well [63,64,65]. The CMZ is also suggested to have contribution in regeneration of the damaged retina in frogs and fish [61, 66]. However, the CMZ seems to be present in the post-hatched bird but not in the adult and absent in mammals (reviewed in ref. 54). Third, the literature indicates that Müller glial cells are the major glial component of the retina, which is one of the last retinal cell types to be born during development [67, 68]. Müller glia are the only cells to span all retinal layers and have processes that contact neighboring neurons and form part of the outer and inner limiting membranes. Thus, Müller glia are well positioned to monitor retinal homeostasis and contribute to retinal structure and function [67]. Mammalian Müller glia have the capacity to locally de-differentiate following retinal injury to become multipotent retinal progenitors, preferentially supplying photoreceptors [69] (reviewed in ref. 70). In this process, Wnt signaling plays a crucial role [71] and this mechanism might share the similar signaling found in fish [43]. Interestingly, the level of photoreceptor cell death-induced Müller glia proliferation is different even among mouse strains (12961/SvJ vs C57BL/6), correlating with the gene expression changes of Cyclin D1 and Nestin [72]. It suggests that the gene regulation could control this damage-induced Müller glia proliferation. Although mammalian Müller glia are still insufficient for fully repairing a damaged retina [67], Müller glial cell-dependent spontaneous regeneration is remarkable in some species such as fish [73]. The mechanisms behind the retina regeneration are also getting more understood in the high regenerative species.
Recent work in Medaka fish showed that a single neurogenic factor, Atoh7, directs Müller glia into proliferation [74]. Atoh7 forces quiescent radial glia into neurons through Notch signaling activation, which is also involved in the maintenance of the glial fate [74, 75]. mTor signaling has been shown to support the formation of proliferating Müller glia progenitor cells in the chick [76]. Comparing the methylation profiles of Müller glia and Müller glia progenitor cell provides DNA methylation landscape during cellular reprogramming and regeneration [77].
Therefore, key to a successful self-repair process in the adult mammalian retina upon injury or degenerative disease is controlling Müller glial regenerative function around in the degenerative regions. Surprisingly, a very recent report showed indeed the mechanisms underlying neuronal regeneration in adult mouse retina [78], suggesting that the regeneration of mammalian retina is not impossible. A promising strategy would be to analyze differences between the transcriptomes and epigenomes of fish and mammalian Müller glia (and Müller glia progenitors) [67, 79]. A recent novel technology, in vivo genome editing, might be an important tool for future approaches to promote a self-repair program in mammalian retina [80] (perhaps by combining with the recent finding that stimulates neuronal regeneration in the retina [78]). A combination of approaches will be needed to probe essential signaling pathways and define those signals that trigger conversion of tissue or specific cells to multipotent retinal progenitors.
In vitro tissue and organ generation
Multicellular organisms have been shown to have a self-organizing ability at the cellular level. Plant calluses, for example, are formed by de-differentiated cells that have tissue-forming ability accompanied by organogenesis [81,82,83]. In animals, dissociated primitive multicellular sponge cells are able to grow and differentiate into a whole sponge [84]. Single-cell suspension culture can reconstitute chick organs [85]. Tumor-derived multi-potent stem cells have tissue-forming potential and these tissues are called embryoid bodies [86]. A single human keratinocyte can generate a stratified squamous epithelium where proliferating and differentiating cells are in a basal layer and upper layer, respectively [87, 88]. The development of growth media has been the basic principle of in vitro tissue culture. Historical aspects of cell, tissue and organoid culture, and their potential applications have been extensively described by Lancaster and Knoblich [89], and Clevers [90].
Organoids are in vitro 3D tissues and organs derived from stem cells or parts of tissues, which have bipotent, multipotent, or pluripotent cells. They also show self-renewal and differentiation capacities in response to a culture environment. Organoid formation is mostly driven by self-organization in a minimal growth factor condition. The organoids typically consist of more than two different cell types and exhibit morphogenetic features closely mimicking an equivalent tissue and organ in vivo. Studies using in vitro systems tend to benefit from an abundant source, production, and easier manipulation.
ESCs can be derived from the inner cell mass of blastocysts, and iPSCs [91, 92] can be reprogramed from dermal fibroblasts [93], blood cells [94, 95], RPE [96], and photoreceptor cells [97, 98] using reprogramming factors (Yamanaka factors) (Fig. 1). Because the inner cell mass generates three germ layers—ectoderm, mesoderm, and endoderm—though gastrulation, its in vitro derivative also shows three-germ-layer differentiation (Fig. 1a, b). iPSCs show a similar extent of differentiation ability to ESCs [99].
These new stem cell technologies facilitate several lines of research related to regenerative medicine. For example, transplantation of ESC- and iPSC-derived retina has been shown to restore vision in rd1 end-stage retinal degeneration mice [100]. A clinical trial is underway, in which human ESC-derived RPE is transplanted into patients with age-related macular degeneration (RPE degeneration leading to photoreceptor loss) [101,102,103]. An advanced personal medicine technique is being tested, involving autologous transplantation of RPE from iPSCs that were generated from patient skin [104].
Repairing damaged heart from the limited regenerative capacity of the adult mammal is a major challenge in medicine. For this purpose, the use of stem cells would be helpful for understanding and enhancing cardiac regeneration [105, 106]. iPSC-derived multi-layered cardiovascular cell sheets that consist of cardiomyocytes, endothelial cells, and vascular mural cells, showed long-term survival of the cell graft to the heart in vivo [107]. Artificial manipulation of cell composition and geometry brings an ideal framework for scale-up to pre-clinical studies [108]. These results suggest that tissue engineering is a promising approach in regenerative medicine [109,110,111].
There are a wide variety of applications for stem cell technologies; infectious disease models using Helicobacter pylori and Zika virus [112, 113], a congenital disease model, microcephaly [114], and an acquired disease model, cancer [115]; Toxicology models such as liver organoid testing hepatocyte function act as a gold standard [116,117,118]. These remarkable advances hold hope for the future of gene therapy and regenerative medicine as more personalized treatments.
Another aspect of stem cell research useful in fundamental research is that it reflects species-specific differences such as organ size, which are hard to analyze via two-dimensional culture systems [112, 114, 119,120,121]. Unique features of the human brain such as the inner fiber layer, outer subventricular zone, and outer radial glia [122,123,124,125], can now be generated in vitro [114]. The period of differentiation in the human brain is longer than that in the mouse [114, 126,127,128]. These human-specific features likely contribute to human brain-specific identity and functions [129]. Advances in this line of research would be supported by genome-editing technologies as powerful tools for basic and biomedical research [130, 131].
We have been trying to develop an ESC-derived brain and retina organoid system in 3D culture using mouse and human embryonic stem cells, asking what are the minimum or essential factors during several phases of brain development. To date, there are several successful methods for generating cerebral cortex, neuroretina, hypothalamus, and adenohypophysis from mouse embryonic stem cells [126, 128, 132,133,134,135]. After we developed mouse ESC methods, further efforts substantially advanced the 3D technologies that enable us to make human brain parts, including retina, in a dish [119, 127, 136, 137]. A key to developing in vitro organogenesis methods is visualizing key marker gene expression in living condition. To do this, researchers typically design a fluorescent reporter system, which is knocked-in at the specific gene loci by genome-editing tools, ZFN, TALEN, and CRISPR/Cas9, resulting in fluorescent cell lines such as Lim3-Venus (pituitary primordium marker), RX-Venus (neuroretina marker), CRX-Venus (photoreceptor progenitor marker), FOXG1-Venus (telencephalic progenitor marker), PAX6-Venus (cells in cortical ventricular zone), Fgf5-mTurquoise2 (epiblast marker), Six3-Tomato (early rostral brain marker), and Irx3-Venus (early caudal brain marker) [119, 127, 135, 138, 139]. This knock-in reporter approach would facilitate performing real-time monitoring including lineage tracing [119], enrichment of specific cell populations, and identification of the reporter-expressing cells when transplanted in vivo in order to rescue hypopituitarism, endocrinological disorders [135]. Using RX-Venus and CRX-Venus human ESCs, a ciliary margin-like stem cell niche was found in optic cups culture [65]. A few other examples showing fluorescent reporter knock-in in pluripotent stem cells have also been described [140,141,142,143,144] (also, see a recent report, showing a 2A-peptide-based knock-in strategy for human iPSC [145]). The genome editing-assisted reporter knock-in strategy might support the identification of specific cell types responsible for self-repair and regeneration.
Because of its simple but remarkable window into self-organization, the 3D method approach is not only providing benefits for regenerative medicine but also modeling organ formation from a new angle in which the intrinsic aspects of organ development—self-assembly, self-patterning and self-morphogenesis—can be observed [146, 147]. For instance, successful live-imaging and live-manipulation of in vitro eye organogenesis allows us to find essential factors involved in the processes [133]. Similar visualization of in vivo processes is extremely difficult in mammals.
Following is a list of recent findings related to the formation of eye organoids (Fig. 1c). Using a diencephalic tissue culture condition, we showed that optic vesicle specification requires Wnt signaling [148]. Using an atomic force microscope and laser-ablation techniques, we demonstrated that relaxation of retina is a critical step for optic cup morphogenesis [133, 149]. By temporally and locally manipulating the Wnt gradient formation, we found that a dorsal–ventral retinal pattern self-forms [150]. Genome-wide analysis and bioinformatics analysis indicates transcriptional profiling of retina and RPE using a transdifferentiation assay [151]. We also provided the first example of a freezing-mediated tissue-storage model using ZFN modified-reporter knock-in human ESCs [119].
Organoids derived from adult stem cells
The adult body has several different tissue (organ)-specific ASCs, which retain the ability to self-renew and replenish all of the cell types as seen in their intact condition through a damage-repair process [152]. Understanding and manipulation of adult stem cells are quite essential for the future of basic and clinical studies in terms of developmental and regenerative aspects.
New technologies allow us to recapitulate organ formation in vitro from ASCs, as their identities are similar to the organ from which they were derived (Fig. 1b). ASC-derived organoids appear to be genetically stable even when cultured over long periods of time [152]. Like pluripotent stem cells, the genomic stability issue is considerably important when researchers try to use ASCs in in vitro culture. Keeping genomic information intact is essential for organ regeneration and to be a safe cell source for regenerative medicine [3].
There have been numerous reports that several ASC-derived organoids can be generated in vitro (reviewed in ref. 152): liver [153, 154], pancreas [155, 156], stomach [157, 158], fallopian tube [159], prostate [160, 161], taste buds [162], and salivary gland [163].
Notably, ASCs require a specific culture environment to mimic their original environment supporting growth and stemness [3]. Even more important is the role of ASCs in regeneration upon damage. They are quiescent until needed and there are biopotent (liver or skin etc) and multipotent (intestinel, stomach, mammary stem cells, hair follicle, and so on) cell states, which are dependent on individual cell types (reviewed in ref. 164). Controlling these essential aspects of ASCs in vitro promises the success of translational research [3]. For instance, organoids can be grafted onto injured intestinal regions, and liver organoids differentiate into functional hepatocytes upon transplantation [3].
Thus, the ASC organoid culture system is a useful tool for genetic diseases, host–pathogen interactions and cancer progression models [152]. Furthermore, the combination of ASC-organotypic culture and CRISPR/Cas9-based gene editing would be obviously powerful; genome-editing technologies appear to be useful particularly in genome level manipulation [155, 165]. An initial attempt was performed using intestinal organoids that contained a patient-derived mutation [166] and subsequent work showed a sequential cancer progression by introducing oncogene-related genes [115]. Some of the ASC-derived organoids have been shown to treat the disease portion by transplantation [167,168,169] (reviewed in refs. 152, 170). Thereby, lineage-restricted stem cells would be expected to be useful for efficient and rapid production of tissues and organs. In this way studying in vitro models that recapitulate human development and pathologies is essential for promoting our understanding of the pathogenesis of diseases, which can lead to the discovery of novel therapeutic paradigms. Genome-editing tools would further widen the potential of ASC technologies.
Vasculature-associated organoid assembles
To support organ growth and homeostasis, the vasculature system performs indispensable roles in vivo. Because organoids show a limited growth potential and maturation level, a main limitation was thought to be the lack of nutrient support from blood vessels. Takebe et al. [171, 172] first demonstrated the creation of a vascularized and functional human liver from human iPSCs by transplantation in an in vivo environment. They succeeded in recapitulating organogenetic interactions between adult tissues, endothelial and mesenchymal cells. This technology highlights liver-specific functions such as protein production and human-specific drug metabolism [171, 172]. This discovery has two important aspects: (1) functional maturation of pluripotent stem-cell-derived tissue requires vascular support and (2) in vivo derived vasculature can be a powerful source of vascular support by connecting with in vitro-derived vasculature. They further strived to obtain a generalized method for organ bud formation from diverse tissues, such as kidney, pancreas, intestine, heart, lung, and brain by carefully modulating mechanical properties of culture environments [173]. However, whether that vasculature could function physiologically as a stem cell niche for complex brain organoids to support growth and maturation remains unknown. In vivo studies show that direct cell–cell contact with the vascular niche is important for neural stem cell function [174,175,176,177]. Therefore, combining several complex in vitro tissues and organs might reproduce better physiological function and provide an advantageous environment for drug screening in regenerative medicine.
Humanized organ generation in vivo
Due to the limitation of availability of donors for organ allotransplantation, exploring other potential resources is essential. In the case of using stem cell technologies, generating organs sufficient for supporting the original function should be considered. It is important that organ size is appropriately fitted for patients. Therefore, to overcome issues with appropriate size, shape and matured functional human-organ requirements upon implantation of organs in the clinical setting is attracting attention. As we mentioned above, even in in vitro conditions, stem cell-derived human organoids reflect species-specific features, such as size and shape. However, complete matured and vascularized human organs are not yet available by in vitro methods. Alternatively, several differences between mice and humans limit the mouse as a model of human disease [178].
Work toward resolving these issues is going to open a new paradigm. It utilizes a chimeric system between closely related species so that cells of a different origin have a less possibility to be rejected in the host environment. Regarding an experimental system of chimerism, there is pioneering work in close species, such as chick–quail, rat–mouse, and sheep–goat [179,180,181,182], and more closely related species, Mus musculus-Mus caroli, M. caroli-M. musculus and Bos taurus–Bos indicus [182, 183]. The successful production of interspecies chimaeras is largely dependent on the matching of the species origin of the trophectoderm derivatives and that of the maternal uterus [184,185,186].
In addition, owing to a dramatic increase in large animal models, mechanisms of pathology and regenerative medicine are becoming more understood and use of those models has become central to new therapeutic strategies [178] and the large animals have been already shown to be engineered (reviewed in refs. 187–189). The domestic pig is one of the most popular animal models for agricultural and biomedical research. Recently, the pig system has enabled researchers to generate several human organs, overcoming not only the number of organs but also organ size (reviewed in ref. 190) (Fig. 2b). In this large animal model, genetically modified chimaeric pigs are used in order to replace pig organs with humanized organs by combining three powerful techniques: generation of interspecies chimaeras, manipulation of pluripotent stem cells, and genome editing. Based on their initial principle established in rodents [191, 192], Nakauchi and colleagues produced a method that potentially allows human PSC-derived organ generation in the vacant space (or an empty developmental niche; see also the interspecies chimerism between mouse and rat [193]) of a non-rodent large animal, the pig [194]. Belmonte and colleagues indeed showed an interspecies chimerism between human and pig using human iPSC, suggesting the possibility of xeno-generating human tissues and organs in the pig [195] (see also a review paper [186]). Pigs with severe combined immunodeficiency (SCID) that were generated by the TALEN system are also available, perhaps providing a useful model for xenotransplantation [196]. There are a couple of potential examples reviewed in detail by Nakauchi of “genetically engineered organ niches” [186, 197].
Interestingly, work showed an example of combining somatic cell nuclear transfer (SCNT) and direct injection of the CRISPR/Cas9 ribonucleoprotein complex which led the generation of a growth-hormone receptor binding protein-10 (GRB10) ablated pig [198]. As such the CRISPR/Cas9 system-mediated genome-modified pigs have also been reported, raising the possibility of their utility for application in regenerative medicine [199, 200]. Although, so far, larger animals such as dogs, pigs, and primates have been used [178, 201, 202], this work suggests that the pig can serve as a better bridge between rodents and primate models in order to study human diseases [198]. This system might provide an unlimited number of humanized organs. Additional examples of genome editing in pigs are available elsewhere [198, 203,204,205,206,207,208].
Conclusions
In the near future, a rapid and reliable method may be developed that allows genetically engineered organ niches in large animal models using genome-editing tools. However, ethical issues need to be carefully considered [209]. In particular, human central nervous system (CNS)-derived organs require special attention. It is quite difficult to use large animal models in order to generate humanized CNS. We can not also exclude the possibility of having new hybrid human–pig viruses in this system. Therefore, transplantation technology without the large animal technology is still a realistic way to replace degenerated portions with autologous patient-derived tissues and organs, which are optionally repaired by genome-editing technology. Alternatively, direct repair of patient degenerative portions by genome-editing technology would be a future challenge (Fig. 2c). To accomplish this ambitious aim, a comprehensive understanding of the essential differences between less regenerative species and highly regenerative species would be needed by taking account of evolutionary changes (or aspects) of organ regeneration. It may provide the basic idea to develop alternative strategies to stimulate self-repair in the low regenerative organs. Stimulating regenerative ability by introducing a genome modification, introducing exogenous cells/tissues (enhancing endogenous regenerative ability), or a combination of both could eventually be possible in less regenerative tissues and organs with the assistance of advanced genome editing tools and stem cell technologies.
References
Eiraku M, Sasai Y. Self-formation of layered neural structures in three-dimensional culture of ES cells. Curr Opin Neurobiol. 2012;22:768–77. https://doi.org/10.1016/j.conb.2012.02.005
Sasai Y. Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell. 2013;12:520–30. https://doi.org/10.1016/j.stem.2013.04.009
Sato T, Clevers H. SnapShot: growing organoids from stem cells. Cell. 2015;161:1700–1700.e1. https://doi.org/10.1016/j.cell.2015.06.028
Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol. 2014;32:773–85. https://doi.org/10.1038/nbt.2958
Song JJ, Ott HC. Organ engineering based on decellularized matrix scaffolds. Trends Mol Med. 2011;17:424–32. https://doi.org/10.1016/j.molmed.2011.03.005
Soto-Gutierrez A, Wertheim JA, Ott HC, Gilbert TW. Perspectives on whole-organ assembly: moving toward transplantation on demand. J Clin Invest. 2012;122:3817–23. https://doi.org/10.1172/Jci61974
Badylak SF, Taylor D, Uygun K. Whole-Organ Tissue Engineering: Decellularization and Recellularization of Three-Dimensional Matrix Scaffolds. Annu Rev Biomed Eng. 2011;13:27–53. https://doi.org/10.1146/annurev-bioeng-071910-124743
Ekser B, Ezzelarab M, Hara H, van der Windt DJ, Wijkstrom M, Bottino R, et al. Clinical xenotransplantation: the next medical revolution? Lancet. 2012;379:672–83. https://doi.org/10.1016/S0140-6736(11)61091-X
Sgroi A, Buhler LH, Morel P, Sykes M, Noel L. International human xenotransplantation inventory. Transplantation. 2010;90:597–603. https://doi.org/10.1097/TP.0b013e3181eb2e8c
Cooper, D. K. A brief history of cross-species organ transplantation. Proc (Bayl Univ Med Cent). 2012;25:49-57.
Peng Y, Clark KJ, Campbell JM, Panetta MR, Guo Y, Ekker SC. Making designer mutants in model organisms. Development. 2014;141:4042–54. https://doi.org/10.1242/dev.102186
Cornu TI, Mussolino C, Cathomen T. Refining strategies to translate genome editing to the clinic. Nat Med. 2017;23:415–23. https://doi.org/10.1038/nm.4313
Blasco RB, Karaca E, Ambrogio C, Cheong TC, Karayol E, Minero VG, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Reports. 2014;9:1219–27. https://doi.org/10.1016/j.celrep.2014.10.051
Li Y, Park AI, Mou H, Colpan C, Bizhanova A, Akama-Garren E, et al. A versatile reporter system for CRISPR-mediated chromosomal rearrangements. Genome Biol. 2015;16:111. https://doi.org/10.1186/s13059-015-0680-7
Jiang J, Zhang L, Zhou X, Chen X, Huang G, Li F, et al. Induction of site-specific chromosomal translocations in embryonic stem cells by CRISPR/Cas9. Sci Rep. 2016;6:21918. https://doi.org/10.1038/srep21918
Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, et al. Editing DNA methylation in the mammalian genome. Cell. 2016;167:233–47.e217. https://doi.org/10.1016/j.cell.2016.08.056
Epigenetics: CRISPR edits gene methylation. Nature 2016;537:588, https://doi.org/10.1038/537588c.
Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–7. https://doi.org/10.1038/nbt.3199
Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods. 2016;13:127–37. https://doi.org/10.1038/nmeth.3733
Xu T, Li Y, Van Nostrand JD, He Z, Zhou J. Cas9-based tools for targeted genome editing and transcriptional control. Appl Environ Microbiol. 2014;80:1544–52. https://doi.org/10.1128/AEM.03786-13
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. https://doi.org/10.1126/science.1225829
Ledford H. CRISPR, the disruptor. Nature. 2015;522:20–4. https://doi.org/10.1038/522020a
Travis J. Making the cut. Science. 2015;350:1456–7. https://doi.org/10.1126/science.350.6267.1456
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide-sequence of the iap gene, responsible for alkaline-phosphatase isozyme conversion in Escherichia coli, and identification of the gene-product. J Bacteriol. 1987;169:5429–33.
Sorek R, Kunin V, Hugenholtz P. CRISPR - a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–6. https://doi.org/10.1038/nrmicro1793
Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–+. https://doi.org/10.1038/nature09523
Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–55. https://doi.org/10.1038/nbt.2842
Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096 https://doi.org/10.1126/science.1258096
DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–43. https://doi.org/10.1093/nar/gkt135
Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv. 2015;33:41–52. https://doi.org/10.1016/j.biotechadv.2014.12.006
Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10:1028–+. https://doi.org/10.1038/Nmeth.2641
Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–8. https://doi.org/10.1016/j.celrep.2013.06.020
Awata H, Watanabe T, Hamanaka Y, Mito T, Noji S, Mizunami M. Knockout crickets for the study of learning and memory: dopamine receptor Dop1 mediates aversive but not appetitive reinforcement in crickets. Sci Rep. 2015;5:15885. https://doi.org/10.1038/srep15885
Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–72. https://doi.org/10.1038/cr.2013.45
Guo X, Zhang T, Hu Z, Zhang Y, Shi Z, Wang Q, et al. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development. 2014;141:707–14. https://doi.org/10.1242/dev.099853
Wang HY, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8. https://doi.org/10.1016/j.cell.2013.04.025
Yang H, Wang HY, Shivalila CS, Cheng AW, Shi LY, Jaenisch R. One-Step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–9. https://doi.org/10.1016/j.cell.2013.08.022
Yang H, Wang HY, Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc. 2014;9:1956–68. https://doi.org/10.1038/nprot.2014.134
Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. https://doi.org/10.1126/science.1231143
Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. https://doi.org/10.1126/science.1232033
Karl MO, Reh TA. Regenerative medicine for retinal diseases: activating endogenous repair mechanisms. Trends Mol Med. 2010;16:193–202. https://doi.org/10.1016/j.molmed.2010.02.003
Ramachandran R, Zhao XF, Goldman D. Insm1a-mediated gene repression is essential for the formation and differentiation of Müller glia-derived progenitors in the injured retina. Nat Cell Biol. 2012;14:1013–+. https://doi.org/10.1038/ncb2586
Wan J, Ramachandran R, Goldman D. HB-EGF is necessary and sufficient for Müller Glia dedifferentiation and retina regeneration. Dev Cell. 2012;22:334–47. https://doi.org/10.1016/j.devcel.2011.11.020
Nagashima M, Barthel LK, Raymond PA. A self-renewing division of zebrafish Müller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development. 2013;140:4510–21. https://doi.org/10.1242/dev.090738
Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JCI. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–9. https://doi.org/10.1038/nature08899
Kikuchi K. Dedifferentiation, transdifferentiation, and proliferation: mechanisms underlying cardiac muscle regeneration in zebrafish. Curr Pathobiol Rep. 2015;3:81–8. https://doi.org/10.1007/s40139-015-0063-5
Kikuchi K, Poss KD. Cardiac regenerative capacity and mechanisms. Annu Rev Cell Dev Biol. 2012;28:719–41. https://doi.org/10.1146/annurev-cellbio-101011-155739
Gemberling M, Bailey TJ, Hyde DR, Poss KD. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013;29:611–20. https://doi.org/10.1016/j.tig.2013.07.003
Hata S, Namae M, Nishina H. Liver development and regeneration: From laboratory study to clinical therapy. Dev Growth Differ. 2007;49:163–70. https://doi.org/10.1111/j.1440-169x.2007.00910.x
Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–80. https://doi.org/10.1126/science.1200708
Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–6.
Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, et al. Modulation of tissue repair by regeneration enhancer elements. Nature. 2016;532:201–6. https://doi.org/10.1038/nature17644
Seeger T, Porteus M, Wu JC. Genome editing in cardiovascular biology. Circ Res. 2017;120:778–80. https://doi.org/10.1161/CIRCRESAHA.116.310197
Ail D, Perron M. Retinal degeneration and regeneration-lessons from fishes and amphibians. Curr Pathobiol Rep. 2017;5:67–78. https://doi.org/10.1007/s40139-017-0127-9
Locker M, Borday C, Perron M. Stemness or not stemness? Current status and perspectives of adult retinal stem cells. Curr Stem Cell Res Ther. 2009;4:118–30.
Lamba D, Karl M, Reh T. Neural regeneration and cell replacement: a view from the eye. Cell Stem Cell. 2008;2:538–49. https://doi.org/10.1016/j.stem.2008.05.002
Stone LS, Steinitz H. Regeneration of neural retina and lens from retina pigment cell grafts in adult newts. J Exp Zool. 1957;135:301–17.
Moshiri A, Close J, Reh TA. Retinal stem cells and regeneration. Int J Dev Biol. 2004;48:1003–14. https://doi.org/10.1387/ijdb.041870am
Yasumuro H, Sakurai K, Toyama F, Maruo F, Chiba C. Implications of a multi-step trigger of retinal regeneration in the adult Newt. Biomedicines. 2017;5. https://doi.org/10.3390/biomedicines5020025.
Merrell AJ, Stanger BZ. Adult cell plasticity in vivo: de-differentiation and transdifferentiation are back in style. Nat Rev Mol Cell Biol. 2016;17:413–25. https://doi.org/10.1038/nrm.2016.24
Fischer AJ, Bosse JL, El-Hodiri HM. The ciliary marginal zone (CMZ) in development and regeneration of the vertebrate eye. Exp Eye Res. 2013;116:199–204. https://doi.org/10.1016/j.exer.2013.08.018
Centanin L, Hoeckendorf B, Wittbrodt J. Fate restriction and multipotency in retinal stem cells. Cell Stem Cell. 2011;9:553–62. https://doi.org/10.1016/j.stem.2011.11.004
Tropepe V, Coles BL, Chiasson BJ, Horsford DJ, Elia AJ, McInnes RR, et al. Retinal stem cells in the adult mammalian eye. Science. 2000;287:2032–6.
Coles BL, Angenieux B, Inoue T, Del Rio-Tsonis K, Spence JR, McInnes RR, et al. Facile isolation and the characterization of human retinal stem cells. Proc Natl Acad Sci USA. 2004;101:15772–7. https://doi.org/10.1073/pnas.0401596101
Kuwahara A, Ozone C, Nakano T, Saito K, Eiraku M, Sasai Y. Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat Commun. 2015;6:6286 https://doi.org/10.1038/ncomms7286
Mitashov VI. Retinal regeneration in amphibians. Int J Dev Biol. 1997;41:893–905.
Goldman D. Müller glial cell reprogramming and retina regeneration. Nat Rev Neurosci. 2014;15:431–42. https://doi.org/10.1038/nrn3723
Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. https://doi.org/10.1002/ar.1092120215
Ooto S, Akagi T, Kageyama R, Akita J, Mandai M, Honda Y, et al. Potential for neural regeneration after neurotoxic injury in the adult mammalian retina. Proc Natl Acad Sci USA. 2004;101:13654–9. https://doi.org/10.1073/pnas.0402129101
Chohan A, Singh U, Kumar A, Kaur J. Müller stem cell dependent retinal regeneration. Clin Chim Acta. 2017;464:160–4. https://doi.org/10.1016/j.cca.2016.11.030
Osakada F, Ooto S, Akagi T, Mandai M, Akaike A, Takahashi M. Wnt signaling promotes regeneration in the retina of adult mammals. J Neurosci. 2007;27:4210–9. https://doi.org/10.1523/JNEUROSCI.4193-06.2007
Suga A, Sadamoto K, Fujii M, Mandai M, Takahashi M. Proliferation potential of Müller glia after retinal damage varies between mouse strains. PLoS ONE. 2014;9:e94556. https://doi.org/10.1371/journal.pone.0094556
Hamon A, Roger JE, Yang XJ, Perron M. Müller glial cell-dependent regeneration of the neural retina: an overview across vertebrate model systems. Dev Dyn. 2016;245:727–38. https://doi.org/10.1002/dvdy.24375
Lust K, Sinn R, Perez Saturnino A, Centanin L, Wittbrodt J. De novo neurogenesis by targeted expression of atoh7 to Müller glia cells. Development. 2016;143:1874–83. https://doi.org/10.1242/dev.135905
Nelson BR, Ueki Y, Reardon S, Karl MO, Georgi S, Hartman BH et al. Genome-wide analysis of Müller glial differentiation reveals a requirement for notch signaling in postmitotic cells to maintain the glial Fate. PLoS ONE. 2011;6. https://doi.org/10.1371/journal.pone.0022817
Zelinka CP, Volkov L, Goodman ZA, Todd L, Palazzo I, Bishop WA, et al. mTor signaling is required for the formation of proliferating Müller glia-derived progenitor cells in the chick retina. Development. 2016;143:1859–73. https://doi.org/10.1242/dev.133215
Powell C, Grant AR, Cornblath E, Goldman D. Analysis of DNA methylation reveals a partial reprogramming of the Müller glia genome during retina regeneration. Proc Natl Acad Sci USA. 2013;110:19814–9. https://doi.org/10.1073/pnas.1312009110
Jorstad NL, Wilken MS, Grimes WN, Wohl SG, VandenBosch LS, Yoshimatsu T, et al. Stimulation of functional neuronal regeneration from Müller glia in adult mice. Nature. 2017;548:103–7. https://doi.org/10.1038/nature23283
Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, et al. The transcriptome of retinal Müller glial cells. J Comp Neurol. 2008;509:225–38. https://doi.org/10.1002/cne.21730
Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–9. https://doi.org/10.1038/nature20565
Krikorian AD, Berquam DL. Plant cell and tissue cultures - role of Haberlandt. Bot Rev. 1969;35:59–+. https://doi.org/10.1007/Bf02859888
Thorpe TA. History of plant tissue culture. Mol Biotechnol. 2007;37:169–80.
Sugiyama M. Historical review of research on plant cell dedifferentiation. J Plant Res. 2015;128:349–59. https://doi.org/10.1007/s10265-015-0706-y
Wilson HV. A new method by which sponges may be artificially reared. Science. 1907;25:912–5. https://doi.org/10.1126/science.25.649.912
Weiss P, Taylor AC. Reconstitution of complete organs from single-cell suspensions of chick embryos in advanced stages of differentiation. Proc Natl Acad Sci USA. 1960;46:1177–85.
Pierce GB Jr, Dixon FJ Jr, Verney EL. Teratocarcinogenic and tissue-forming potentials of the cell types comprising neoplastic embryoid bodies. Lab Invest. 1960;9:583–602.
Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–43.
Rheinwald JG, Green H. Formation of a keratinizing epithelium in culture by a cloned cell line derived from a teratoma. Cell. 1975;6:317–30.
Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345. https://doi.org/10.1126/science.1247125
Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–97. https://doi.org/10.1016/j.cell.2016.05.082
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. https://doi.org/10.1016/j.cell.2006.07.024
Shi Y, Inoue H, Wu JC, Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov. 2017;16:115–30. https://doi.org/10.1038/nrd.2016.245
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. https://doi.org/10.1016/j.cell.2007.11.019
Zhou H, Martinez H, Sun B, Li A, Zimmer M, Katsanis N, et al. Rapid and efficient generation of transgene-free iPSC from a small volume of cryopreserved blood. Stem Cell Rev. 2015;11:652–65. https://doi.org/10.1007/s12015-015-9586-8
Agu CA, Soares FAC, Alderton A, Patel M, Ansari R, Patel S, et al. Successful generation of human induced pluripotent stem cell lines from blood samples held at room temperature for up to 48 hr. Stem Cell Rep. 2015;5:660–71. https://doi.org/10.1016/j.stemcr.2015.08.012
Hu Q, Friedrich AM, Johnson LV, Clegg DO. Memory in induced pluripotent stem cells: reprogrammed human retinal-pigmented epithelial cells show tendency for spontaneous redifferentiation. Stem Cells. 2010;28:1981–91. https://doi.org/10.1002/stem.531
Hiler D, Chen X, Hazen J, Kupriyanov S, Carroll PA, Qu C, et al. Quantification of Retinogenesis in 3D cultures reveals epigenetic memory and higher efficiency in iPSCs derived from rod photoreceptors. Cell Stem Cell. 2015;17:101–15. https://doi.org/10.1016/j.stem.2015.05.015
Hiler DJ, Barabas ME, Griffiths LM, Dyer MA. Reprogramming of mouse retinal neurons and standardized quantification of their differentiation in 3D retinal cultures. Nat Protoc. 2016;11:1955–76. https://doi.org/10.1038/nprot.2016.109
Yamanaka S. A fresh look at iPS cells. Cell. 2009;137:13–7. https://doi.org/10.1016/j.cell.2009.03.034
Mandai M, Fujii M, Hashiguchi T, Sunagawa GA, Ito S, Sun JN, et al. iPSC-derived retina transplants improve vision in rd1 end-stage retinal-degeneration mice (vol 8, pg 69, 2017). Stem Cell Rep. 2017;8:1112–3. https://doi.org/10.1016/j.stemcr.2017.03.024
Schwartz SD, Regillo CD, Lam BL, Eliott D, Rosenfeld PJ, Gregori NZ, et al. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet. 2015;385:509–16. https://doi.org/10.1016/S0140-6736(14)61376-3
Schwartz SD, Tan G, Hosseini H, Nagiel A. Subretinal transplantation of embryonic stem cell-derived retinal pigment epithelium for the treatment of macular degeneration: an assessment at 4 years. Invest Ophth Vis Sci. 2016;57. https://doi.org/10.1167/iovs.15-18681.
Chiba C. The retinal pigment epithelium: an important player of retinal disorders and regeneration. Exp Eye Res. 2014;123:107–14. https://doi.org/10.1016/j.exer.2013.07.009
Mandai M, Watanabe A, Kurimoto Y, Hirami Y, Morinaga C, Daimon T, et al. Autologous induced stem-cell-derived retinal cells for macular degeneration. N Engl J Med. 2017;376:1038–46. https://doi.org/10.1056/NEJMoa1608368
Galdos FX, Guo YX, Paige SL, VanDusen NJ, Wu SM, Pu WT. Cardiac regeneration lessons from development. Circ Res. 2017;120:941–59. https://doi.org/10.1161/Circresaha.116.309040
Doppler SA, Deutsch MA, Serpooshan V, Li G, Dzilic E, Lange R, et al. Mammalian heart regeneration the race to the finish line. Circ Res. 2017;120:630–2. https://doi.org/10.1161/Circresaha.116.310051
Matsuo T, Masumoto H, Tajima S, Ikuno T, Katayama S, Minakata K, et al. Efficient long-term survival of cell grafts after myocardial infarction with thick viable cardiac tissue entirely from pluripotent stem cells. Sci Rep. 2015;5:16842. https://doi.org/10.1038/srep16842
Nakane T, Masumoto H, Tinney JP, Yuan F, Kowalski WJ, Ye F, et al. Impact of cell composition and geometry on human induced pluripotent stem cells-derived engineered cardiac tissue. Sci Rep. 2017;7:45641. https://doi.org/10.1038/srep45641
Yamashita JK. ES and iPS cell research for cardiovascular regeneration. Exp Cell Res. 2010;316:2555–9. https://doi.org/10.1016/j.yexcr.2010.04.004
Masumoto H, Yamashita JK. Human iPS cell-derived cardiac tissue sheets: a platform for cardiac regeneration. Curr Treat Options Cardiovasc Med. 2016;18:65 https://doi.org/10.1007/s11936-016-0489-z
Shkumatov A, Baek K, Kong H. Matrix rigidity-modulated cardiovascular organoid formation from embryoid bodies. PLoS ONE. 2014;9:e94764. https://doi.org/10.1371/journal.pone.0094764
McCracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–4. https://doi.org/10.1038/nature13863
Qian XY, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–54. https://doi.org/10.1016/j.cell.2016.04.032
Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–+. https://doi.org/10.1038/nature12517
Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–7. https://doi.org/10.1038/nature14415
Soldatow VY, Lecluyse EL, Griffith LG, Rusyn I. In vitro models for liver toxicity testing. Toxicol Res. 2013;2:23–39. https://doi.org/10.1039/C2TX20051A
Nantasanti S, de Bruin A, Rothuizen J, Penning LC, Schotanus BA. Concise review: organoids are a powerful tool for the study of liver disease and personalized treatment design in humans and animals. Stem Cells Transl Med. 2016;5:325–30. https://doi.org/10.5966/sctm.2015-0152
Avior Y, Sagi I, Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat Rev Mol Cell Biol. 2016;17:170–82. https://doi.org/10.1038/nrm.2015.27
Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–85. https://doi.org/10.1016/j.stem.2012.05.009
Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–8. https://doi.org/10.1038/nature15695
McCauley HA, Wells JM. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development. 2017;144:958–62. https://doi.org/10.1242/dev.140731
Fietz SA, Kelava I, Vogt J, Wilsch-Brauninger M, Stenzel D, Fish JL, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–9. https://doi.org/10.1038/nn.2553
Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–61. https://doi.org/10.1038/nature08845
Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex. 2002;12:37–53.
Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–95. https://doi.org/10.1523/JNEUROSCI.4773-10.2011
Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–32. https://doi.org/10.1016/j.stem.2008.09.002
Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci USA. 2013;110:20284–9. https://doi.org/10.1073/pnas.1315710110
Nasu M, Takata N, Danjo T, Sakaguchi H, Kadoshima T, Futaki S, et al. Robust formation and maintenance of continuous stratified cortical neuroepithelium by laminin-containing matrix in mouse ES cell culture. PLoS ONE. 2012;7:e53024. https://doi.org/10.1371/journal.pone.0053024
Kelava I, Lancaster MA. Dishing out mini-brains: current progress and future prospects in brain organoid research. Dev Biol. 2016;420:199–209. https://doi.org/10.1016/j.ydbio.2016.06.037
Savic N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl Res. 2016;168:15–21. https://doi.org/10.1016/j.trsl.2015.09.008
Hockemeyer D, Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell. 2016;18:573–86. https://doi.org/10.1016/j.stem.2016.04.013
Wataya T, Ando S, Muguruma K, Ikeda H, Watanabe K, Eiraku M, et al. Minimization of exogenous signals in ES cell culture induces rostral hypothalamic differentiation. Proc Natl Acad Sci USA. 2008;105:11796–801. https://doi.org/10.1073/pnas.0803078105
Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–6. https://doi.org/10.1038/nature09941
Sasai Y. Grow your own eye: biologists have coaxed cells to form a retina, a step toward growing replacement organs outside the body. Sci Am. 2012;307:44–9.
Suga H, Kadoshima T, Minaguchi M, Ohgushi M, Soen M, Nakano T, et al. Self-formation of functional adenohypophysis in three-dimensional culture. Nature. 2011;480:57–62. https://doi.org/10.1038/nature10637
Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, et al. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun. 2015;6:8896 https://doi.org/10.1038/ncomms9896
Ozone C, Suga H, Eiraku M, Kadoshima T, Yonemura S, Takata N, et al. Functional anterior pituitary generated in self-organizing culture of human embryonic stem cells. Nat Commun. 2016;7:10351. https://doi.org/10.1038/ncomms10351
Takata N, Sakakura E, Kasukawa T, Sakuma T, Yamamoto T, Sasai Y. Establishment of functional genomics pipeline in mouse epiblast-like tissue by combining transcriptomic analysis and gene knockdown/knockin/knockout, using RNA interference and CRISPR/Cas9. Hum Gene Ther. 2016;27:436–50. https://doi.org/10.1089/hum.2015.148
Takata N, Sakakura E, Sasai Y. IGF-2/IGF-1R signaling has distinct effects on Sox1, Irx3, and Six3 expressions during ES cell derived-neuroectoderm development in vitro. In Vitro Cell Dev Biol Anim. 2016;52:607–15. https://doi.org/10.1007/s11626-016-0012-6
Nie J, Hashino E. Organoid technologies meet genome engineering. EMBO Rep. 2017;18:367–76. https://doi.org/10.15252/embr.201643732
Brookhouser N, Raman S, Potts C, Brafman DA. May I cut in? gene editing approaches in human induced pluripotent stem cells. Cells. 2017;6. https://doi.org/10.3390/cells6010005.
Verma N, Zhu Z, Huangfu D. CRISPR/cas-mediated knockin in human pluripotent stem cells. Methods Mol Biol. 2017;1513:119–40. https://doi.org/10.1007/978-1-4939-6539-7_9
Blair JD, Bateup HS, Hockemeyer DF. Establishment of genome-edited human pluripotent stem cell lines: from targeting to isolation. J Vis Exp. 2016;e53583. https://doi.org/10.3791/53583.
Sluch VM, Davis CHO, Ranganathan V, Kerr JM, Krick K, Martin R et al. Differentiation of human ESCs to retinal ganglion cells using a CRISPR engineered reporter cell line. Sci Rep. 2015;5. https://doi.org/10.1038/srep16595
Homma K, Usui S, Kaneda M. Knock-in strategy at 3’-end of Crx gene by CRISPR/Cas9 system shows the gene expression profiles during human photoreceptor differentiation. Genes Cells. 2017;22:250–64. https://doi.org/10.1111/gtc.12472
Sasai Y. Cytosystems dynamics in self-organization of tissue architecture. Nature. 2013;493:318–26. https://doi.org/10.1038/nature11859
Sasai Y, Eiraku M, Suga H. In vitro organogenesis in three dimensions: self-organising stem cells. Development. 2012;139:4111–21. https://doi.org/10.1242/dev.079590
Sakakura E, Eiraku M, Takata N. Specification of embryonic stem cell-derived tissues into eye fields by Wnt signaling using rostral diencephalic tissue-inducing culture. Mech Develop. 2016;141:90–9. https://doi.org/10.1016/j.mod.2016.05.001
Eiraku M, Adachi T, Sasai Y. Relaxation-expansion model for self-driven retinal morphogenesis: a hypothesis from the perspective of biosystems dynamics at the multi-cellular level. Bioessays. 2012;34:17–25. https://doi.org/10.1002/bies.201100070
Hasegawa Y, Takata N, Okuda S, Kawada M, Eiraku M, Sasai Y. Emergence of dorsal-ventral polarity in ESC-derived retinal tissue. Development. 2016;143:3895–906. https://doi.org/10.1242/dev.134601
Andrabi M, Kuraku S, Takata N, Sasai Y, Love NR. Comparative, transcriptome analysis of self-organizing optic tissues. Sci Data. 2015;2. https://doi.org/10.1038/sdata.2015.30.
Drost J, Clevers H. Translational applications of adult stem cell-derived organoids. Development. 2017;144:968–75. https://doi.org/10.1242/dev.140566
Huch M, Boj SF, Clevers H. Lgr5(+) liver stem cells, hepatic organoids and regenerative medicine. Regen Med. 2013;8:385–7. https://doi.org/10.2217/rme.13.39
Huch M, Koo BK. Modeling mouse and human development using organoid cultures. Development. 2015;142:3113–25. https://doi.org/10.1242/dev.118570
Boj SF, Hwang CI, Baker LA, Chio IIC, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic. Cancer Cell. 2015;160:324–38. https://doi.org/10.1016/j.cell.2014.12.021
Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJM, van de Wetering M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708–21. https://doi.org/10.1038/emboj.2013.204
Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. https://doi.org/10.1016/j.stem.2009.11.013
Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–136.e6. https://doi.org/10.1053/j.gastro.2014.09.042
Kessler M, Hoffmann K, Brinkmann V, Thieck O, Jackisch S, Toelle B et al. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat Commun. 2015;6. https://doi.org/10.1038/ncomms9989
Chua CW, Shibata M, Lei M, Toivanen R, Barlow LJ, Bergren SK, et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat Cell Biol. 2014;16:951–61. https://doi.org/10.1038/ncb3047
Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–75. https://doi.org/10.1016/j.cell.2014.08.017
Ren W, Lewandowski BC, Watson J, Aihara E, Iwatsuki K, Bachmanov AA, et al. Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc Natl Acad Sci USA. 2014;111:16401–6. https://doi.org/10.1073/pnas.1409064111
Maimets M, Rocchi C, Bron R, Pringle S, Kuipers J, Giepmans BN, et al. Long-term in vitro expansion of salivary gland stem cells driven by Wnt signals. Stem Cell Rep. 2016;6:150–62. https://doi.org/10.1016/j.stemcr.2015.11.009
Clevers H. What is an adult stem cell? Science. 2015;350:1319–20. https://doi.org/10.1126/science.aad7016
Dutta D, Heo I, Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med. 2017. https://doi.org/10.1016/j.molmed.2017.02.007.
Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–8. https://doi.org/10.1016/j.stem.2013.11.002
Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat Med. 2012;18:618–23. https://doi.org/10.1038/nm.2695
Fordham RP, Yui S, Hannan NR, Soendergaard C, Madgwick A, Schweiger PJ, et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–44. https://doi.org/10.1016/j.stem.2013.09.015
Fukuda M, Mizutani T, Mochizuki W, Matsumoto T, Nozaki K, Sakamaki Y, et al. Small intestinal stem cell identity is maintained with functional Paneth cells in heterotopically grafted epithelium onto the colon. Gene Dev. 2014;28:1752–7. https://doi.org/10.1101/gad.245233.114
Nakamura T, Watanabe M. Intestinal stem cell transplantation. J Gastroenterol. 2017;52:151–7. https://doi.org/10.1007/s00535-016-1288-8
Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–4. https://doi.org/10.1038/nature12271
Takebe T, Zhang RR, Koike H, Kimura M, Yoshizawa E, Enomura M, et al. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nat Protoc. 2014;9:396–409. https://doi.org/10.1038/nprot.2014.020
Takebe T, Enomura M, Yoshizawa E, Kimura M, Koike H, Ueno Y, et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell. 2015;16:556–65. https://doi.org/10.1016/j.stem.2015.03.004
Ottone C, Krusche B, Whitby A, Clements M, Quadrato G, Pitulescu ME, et al. Direct cell-cell contact with the vascular niche maintains quiescent neural stem cells. Nat Cell Biol. 2014;16:1045–+. https://doi.org/10.1038/ncb3045
Goldberg JS, Hirschi KK. Diverse roles of the vasculature within the neural stem cell niche. Regen Med. 2009;4:879–97. https://doi.org/10.2217/Rme.09.61
Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, et al. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279–88. https://doi.org/10.1016/j.stem.2008.07.025
Rafii S, Butler JM, Ding BS. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529:316–25. https://doi.org/10.1038/nature17040
Whitelaw CB, Sheets TP, Lillico SG, Telugu BP. Engineering large animal models of human disease. J Pathol. 2016;238:247–56. https://doi.org/10.1002/path.4648
Le Douarin NM. The ontogeny of the neural crest in avian embryo chimaeras. Nature. 1980;286:663–9.
Gardner RL, Johnson MH. Investigation of early mammalian development using interspecific chimaeras between rat and mouse. Nat New Biol. 1973;246:86–9.
Fehilly CB, Willadsen SM, Tucker EM. Interspecific chimaerism between sheep and goat. Nature. 1984;307:634–6. https://doi.org/10.1038/307634a0
Williams TJ, Munro RK, Shelton JN. Production of interspecies chimeric calves by aggregation of Bos indicus and Bos taurus demi-embryos. Reprod Fertil Dev. 1990;2:385–94.
Rossant J, Frels WI. Interspecific chimeras in mammals - successful production of live chimeras between mus-musculus and mus-caroli. Science. 1980;208:419–21. https://doi.org/10.1126/science.7367871
Rossant J, Croy BA, Chapman VM, Siracusa L, Clark DA. Interspecific chimeras in mammals: a new experimental system. J Anim Sci. 1982;55:1241–8.
Rossant J, Mauro VM, Croy BA. Importance of trophoblast genotype for survival of interspecific murine chimaeras. J Embryol Exp Morphol. 1982;69:141–9.
Suchy F, Nakauchi H. Lessons from interspecies mammalian chimeras. Annu Rev Cell Dev Biol. 2017. https://doi.org/10.1146/annurev-cellbio-100616-060654.
Aigner B, Renner S, Kessler B, Klymiuk N, Kurome M, Wunsch A, et al. Transgenic pigs as models for translational biomedical research. J Mol Med. 2010;88:653–64. https://doi.org/10.1007/s00109-010-0610-9
Prather RS, Lorson M, Ross JW, Whyte JJ, Walters E. Genetically engineered pig models for human diseases. Annu Rev Anim Biosci. 2013;1:203–19. https://doi.org/10.1146/annurev-animal-031412-103715
Flisikowska T, Kind A, Schnieke A. Genetically modified pigs to model human diseases. J Appl Genet. 2014;55:53–64. https://doi.org/10.1007/s13353-013-0182-9
Feng W, Dai Y, Mou L, Cooper DK, Shi D, Cai Z. The potential of the combination of CRISPR/Cas9 and pluripotent stem cells to provide human organs from chimaeric pigs. Int J Mol Sci. 2015;16:6545–56. https://doi.org/10.3390/ijms16036545
Kobayashi T, Yamaguchi T, Hamanaka S, Kato-Itoh M, Yamazaki Y, Ibata M, et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell. 2010;142:787–99. https://doi.org/10.1016/j.cell.2010.07.039
Usui J, Kobayashi T, Yamaguchi T, Knisely AS, Nishinakamura R, Nakauchi H. Generation of kidney from pluripotent stem cells via blastocyst complementation. Am J Pathol. 2012;180:2417–26. https://doi.org/10.1016/j.ajpath.2012.03.007
Yamaguchi T, Sato H, Kato-Itoh M, Goto T, Hara H, Sanbo M, et al. Interspecies organogenesis generates autologous functional islets. Nature. 2017;542:191–6. https://doi.org/10.1038/nature21070
Matsunari H, Nagashima H, Watanabe M, Umeyama K, Nakano K, Nagaya M, et al. Blastocyst complementation generates exogenic pancreas in vivo in apancreatic cloned pigs. Proc Natl Acad Sci USA. 2013;110:4557–62. https://doi.org/10.1073/pnas.1222902110
Wu J, Platero-Luengo A, Sakurai M, Sugawara A, Gil MA, Yamauchi T, et al. Interspecies chimerism with mammalian pluripotent stem cells. Cell. 2017;168:473–486.e415. https://doi.org/10.1016/j.cell.2016.12.036
Lee K, Kwon DN, Ezashi T, Choi YJ, Park C, Ericsson AC, et al. Engraftment of human iPS cells and allogeneic porcine cells into pigs with inactivated RAG2 and accompanying severe combined immunodeficiency. Proc Natl Acad Sci USA. 2014;111:7260–5. https://doi.org/10.1073/pnas.1406376111
Rashid T, Kobayashi T, Nakauchi H. Revisiting the flight of Icarus: making human organs from PSCs with large animal chimeras. Cell Stem Cell. 2014;15:406–9. https://doi.org/10.1016/j.stem.2014.09.013
Sheets TP, Park CH, Park KE, Powell A, Donovan DM, Telugu BP. Somatic cell nuclear transfer followed by CRIPSR/Cas9 microinjection results in highly efficient genome editing in cloned pigs. Int J Mol Sci. 2016;17. https://doi.org/10.3390/ijms17122031.
Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24:372–5. https://doi.org/10.1038/cr.2014.11
Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, et al. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod. 2014;91. https://doi.org/10.1095/biolreprod.114.121723 doi:ARTN 7810.1095/biolreprod.114.121723.
Casal M, Haskins M. Large animal models and gene therapy. Eur J Hum Genet. 2006;14:266–72. https://doi.org/10.1038/sj.ejhg.5201535
Camus S, Ko WK, Pioli E, Bezard E. Why bother using non-human primate models of cognitive disorders in translational research? Neurobiol Learn Mem. 2015;124:123–9. https://doi.org/10.1016/j.nlm.2015.06.012
Yao J, Huang J, Zhao J. Genome editing revolutionize the creation of genetically modified pigs for modeling human diseases. Hum Genet. 2016;135:1093–105. https://doi.org/10.1007/s00439-016-1710-6
Yum SY, Yoon KY, Lee CI, Lee BC, Jang G. Transgenesis for pig models. J Vet Sci. 2016;17:261–8. https://doi.org/10.4142/jvs.2016.17.3.261
Butler JR, Ladowski JM, Martens GR, Tector M, Tector AJ. Recent advances in genome editing and creation of genetically modified pigs. Int J Surg. 2015;23:217–22. https://doi.org/10.1016/j.ijsu.2015.07.684
Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, et al. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci. 2015;72:1175–84. https://doi.org/10.1007/s00018-014-1744-7
Tan WS, Carlson DF, Walton MW, Fahrenkrug SC, Hackett PB. Precision editing of large animal genomes. Adv Genet. 2012;80:37–97. https://doi.org/10.1016/B978-0-12-404742-6.00002-8
Tanihara F, Takemoto T, Kitagawa E, Rao S, Do LT, Onishi A, et al. Somatic cell reprogramming-free generation of genetically modified pigs. Sci Adv. 2016;2:e1600803. https://doi.org/10.1126/sciadv.1600803
Shaw D, Dondorp W, Geijsen N, de Wert G. Creating human organs in chimaera pigs: an ethical source of immunocompatible organs? J Med Ethics. 2015;41:970–4. https://doi.org/10.1136/medethics-2014-102224
Acknowledgements
We thank Dr. G. Oliver for the informative knowledge. We also thank M. Oxendine for his careful English proofreading. This work was supported by JSPS KAKENHI Grant Number 26112723 (to M.E.).
Author information
Authors and Affiliations
Contributions
N.T. prepared the figures and wrote the manuscript. M.E. supervised the figure preparation and the manuscript writing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Takata, N., Eiraku, M. Stem cells and genome editing: approaches to tissue regeneration and regenerative medicine. J Hum Genet 63, 165–178 (2018). https://doi.org/10.1038/s10038-017-0348-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-017-0348-0
This article is cited by
-
Machine learning-based estimation of spatial gene expression pattern during ESC-derived retinal organoid development
Scientific Reports (2023)
-
The evolving definition of salivary gland stem cells
npj Regenerative Medicine (2021)