Abstract
Clarifying allele frequencies of disease-related genetic variants in a population is important in genomic medicine; however, such data is not yet available for the Japanese population. To estimate frequencies of actionable pathogenic variants in the Japanese population, we examined the reported pathological variants in genes recommended by the American College of Medical Genetics and Genomics (ACMG) in our reference panel of genomic variations, 2KJPN, which was created by whole-genome sequencing of 2049 individuals of the resident cohort of the Tohoku Medical Megabank Project. We searched for pathogenic variants in 2KJPN for 57 autosomal ACMG-recommended genes responsible for 26 diseases and then examined their frequencies. By referring to public databases of pathogenic variations, we identified 143 reported pathogenic variants in 2KJPN for the 57 ACMG recommended genes based on a classification system. At the individual level, 21% of the individuals were found to have at least one reported pathogenic allele. We then conducted a literature survey to review the variants and to check for evidence of pathogenicity. Our results suggest that a substantial number of people have reported pathogenic alleles for the ACMG genes, and reviewing variants is indispensable for constructing the information infrastructure of genomic medicine for the Japanese population.
Similar content being viewed by others
Introduction
One of the main goals of medical genomics is the development of personalized medicine and personalized healthcare based on individual genomes. Large-scale sequencing of individual genomes from cohort participants [1,2,3] provides us with a catalog of numerous genomic variants whose frequencies range from rare to common, as well as a set of phased variants in individual haplotypes. Such genomic variation data of local populations are valuable resources. Thus, this data allows for genome-wide association studies aimed at finding disease-related variants by using phased variants for genotype imputation [4]. In addition to binary phenotypic traits, studying association with quantitative traits, such as metabolomics in plasma, is also valuable to reveal genetic susceptibility to proximal phenotypes at the molecular level [5]. Another important use of this data is to allow for detection of rare and pathogenic variants, and to estimate their population frequencies.
Large-scale genome sequencing of volunteers from general residents provides valuable data for the field of medical genomics; however, whole genome sequencing (WGS) or whole exome sequencing (WES) [6, 7] may uncover other clinically relevant variants in addition to specific findings intended by a particular project. Therefore, it raises an important problem: how can researchers use and manage secondary findings, which may be deliberately sought, or incidental findings (accidental discoveries) from WGS or WES studies? In this situation, the American Colleges of Medical Genetics and Genomics (ACMG) recommends that clinical sequencing laboratories return pathogenic variants of 24 conditions (56 genes) in 2013, and recently updated this list to include 26 diseases (59 genes) [8, 9] as a minimum set for returning secondary findings, which were selected from the viewpoint of medical actionability. Although medical actionability depends on the clinical systems in the society, it is important to estimate population frequencies of the genetic variants in the ACMG gene list to improve social welfare.
Since the ACMG released their recommendations, several groups have tried to estimate frequencies of actionable variants in 56 genes for diverse samples and by using different methods [10,11,12,13,14,15]. Using WGS or WES data, some studies [15, 16] tried to estimate the frequencies of pathogenic variants in the recommended genes for European and African ancestries, and target populations of the 1000 Genomes Project. Although East Asian populations were analyzed in the 1000 Genomes Project, the number of individuals for each population was not significant enough to detect rare pathogenic variants and frequency estimations. Therefore, the frequencies of low-frequency pathogenic variants in the Japanese population have not been characterized well.
Tohoku University Tohoku Medical Megabank Organization (ToMMo) initiated genome cohort studies [17] along with Iwate Medical University to promote research in medical genomics aiming to realize personalized healthcare. As the first step towards our goal, deep WGS of more than 2000 cohort participants was performed, and the reference panel for the Japanese population, 2KJPN, was constructed [18, 19]. In this study, we report the first examination of population frequencies of the responsible genomic variants in the actionable ACMG genes by using 2KJPN and public annotations in the Human Gene Mutation Database (HGMD) [20] and ClinVar [21]. We found that 21% of the individuals had at least one reported pathogenic variant for the 57 autosomal ACMG genes, suggesting that not a small proportion of individuals may have some risk allele for the actionable genes. In addition, we performed manual inspections of some variants through extensive literature surveys, and found that there were many discrepancies between the two public annotations. Some reported disease mutations can be benign variants, and a few variants were lacking enough evidence for the Japanese population. These results indicate that we need to construct an information infrastructure of pathogenic variants for the Japanese population through appropriate variant review and interpretations, ultimately allowing for personalized healthcare for the Japanese population.
Materials and Methods
Subjects and data for single nucleotide variation
We used the 2KJPN whole genome reference panel (2049 individuals) of the Tohoku Medical Megabank Project, which was created using the approach same as that used for the 1KJPN panel (1070 individuals) [18]. Briefly, the subjects were selected from the participants of the resident cohort study [17], and then the genomic DNA of the 2049 individuals obtained from peripheral blood samples was subjected to paired-end sequencing using the Illumina HiSeq 2500 platform (see details in Nagasaki et al. [18]).
This project was performed as a part of prospective cohort studies at ToMMo with the approval of the Ethical Committee of the Tohoku University School of Medicine and ToMMo. The samples used here were obtained from the cohort participants, all of whom gave their written consent. Under the terms of the informed consent provided by the participants in our cohort project, whole genome data including sequenced data, variant calls, and inferred genotypes are securely controlled under the Materials and Information Distribution Review Committee of Tohoku Medical Megabank Project, and the sharing of data with other researchers was discussed in each research proposal by the review committee.
There are two sets of variants in 2KJPN—a high confidence variant set and a high sensitivity variant set. The former set was created with high precision and the latter set was created by maximizing sensitivity. The allele frequency of the high confidence variant set is publicly available through a portal site, the integrative Japanese Genome Variation Database (iJGVD; http://ijgvd.megabank.tohoku.ac.jp/) [19]. In this study, we used both the variant sets (high confidence single nucleotide variations (SNVs) and high sensitive SNVs) for analyses, and the results from the high confidence set were primarily used for subsequent manual inspection. The results of the high sensitive set were also used when additional existing SNVs were suspected. We also used variant frequency data for di-allelic SNVs for 4300 European Americans (EAs) and 2203 African Americans (AAs) from the Exome Sequencing Project (ESP) [22] to compare the allele frequency of each SNV with the corresponding SNV in 2KJPN.
Variant annotation
SnpEff software (ver. 3.3c), which is based on the gene annotation model of GENCODE version 17, was used to predict the effects of a variant on its gene product. SNVs were classified into functional categories, such as synonymous, missense, nonsense, intron, 5′-untranslated region (UTR), and 3′-UTR. As a measure to predict pathogenicity, the combined annotation-dependent depletion (CADD) scores [23] were added to each SNV by intersecting the list of the precomputed scores with all possible SNVs. In addition, for low-frequency missense SNVs, the Mendelian Clinically Applicable Pathogenicity (M-CAP) scores [24] were annotated similarly. To identify which SNV is a reported pathogenic variant, we used the Human Gene Mutation Database (HGMD) Professional (2016.2) [20] and ClinVar (2016 September) [21] (Fig. 1). With the ClinVar database, we used entries that included annotations as “pathogenic” or “likely pathogenic.” Overlaps between the SNVs of 2KJPN and the reported pathological variants in HGMD and ClinVar were extracted. Possible pathological SNVs were identified based on the genomic coordinates and the consistency of the allele bases. We identified 6862 pathological SNVs that overlapped with HGMD or ClinVar variants that were annotated as “pathogenic” or “likely pathogenic.” Then we selected the variants for 57 autosomal genes (except for two X-linked genes: GLA and OTC) recommended by ACMG for the return of genomic results [8] with modifications in 2016 [9].
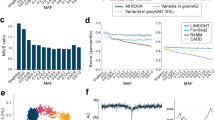
Scheme of analysis pipeline for identifying reported pathogenic variants for the ACMG genes in 2KPN. About 28 M SNVs in 2KJPN were annotated with functional and pathological information by using SnpEff, HGMD, ClinVar, and CADD. Then variants for the 57 autosomal ACMG genes were selected and used for analysis. For comparison with other ethnic populations, allele frequency of autosomal bi-allelic SNVs for EAs (n = 4,300) and AAs (n = 2,203) were used
Filtering variants
To search for disease-causing variants for the 57 autosomal ACMG recommended genes, 2KJPN SNVs that matched the HGMD or ClinVar variants were extracted. Next, we defined the following three categories for these potentially pathogenic variants (Table 1): i) reported pathogenic (RP) variants that were already annotated as “disease-causing mutation (DM)” in HGMD or “pathogenic” in ClinVar; ii) candidates of pathogenic variants (canP) that were already annotated as “DM? (likely disease-causing mutation)” in HGMD or “likely pathogenic” in ClinVar; and iii) disease-associated variants and other types (assoV, etc.). RP and canP variants were filtered by minor allele frequency (MAF) < 0.5% in 2KJPN, and the pathologically annotated SNVs that existed at the higher frequency ( ≥ 0.5%) were classified in the third group (assoV, etc.).
Results
A total of 46,822 SNVs, including 1317 with protein-altering mutations and 386 that were identified in HGMD or ClinVar as “pathogenic” or “likely pathogenic”, in the 57 ACMG genes in 2KJPN were selected based on the genomic coordinates of the genes (see Table 2 for statistics, and a whole list of the variants is shown in Supplementary Table 1). After automatically classifying variants as either RP or canP, 143 SNVs with RP variants were detected with the MAF threshold of < 0.5% (156 RP variants when MAF < 1%) (Table 2). RP or canP variants were found in 47 genes, but not in PSM2, VHL, PTEN, SDHAF2, SDHC, TGFBR1, SMAD3, TNNI3, TPM1, MYL3, ACTC1, PRKAG2, MYL2, DSC2, and AMAD4. Using the allele frequencies of the RP variants, population frequencies of potential risk alleles were estimated (Table 2). Genes that showed relatively higher population frequencies were RYR2, MSH2, MYBPC3, ATP7B, APC, and BRCA2.
At the individual level, 431 of the 2049 individuals had at least one RP variant for the 26 diseases (Fig. 2). This was based on the automatic classification, which may be overestimated in a proportion of individuals having a real risk allele. We then focused on several diseases, and the reported pathogenic variants were manually inspected through a literature survey. We manually reviewed the detected pathogenic variants that have been previously reported by focusing on the distinct phenotypic effects of variants in a single gene (if any), the allele frequencies, and incidence rates. In addition to the pathogenic variants that were previously reported, we also searched for candidates of expected pathogenic variants based on gene-based annotations and predicted scores of pathogenicity. With thresholds of CADD score >20 or M-CAP [24] score >0.025, 815 SNVs (including 709 missense SNVs) were detected as variants which satisfy a recommended threshold of pathogenicity but lack any pathological annotations in HGMD and ClinVar (Supplementary Table 2). For example, we found three nonsense variants and 37 missense variants in apolipoprotein B gene (APOB) that were not pathogenically annotated in HGMD and ClinVar. The three nonsense variants of APOB were p.Tyr1578*, p.Ser2128*, and p.Lys2376*, and were all found as singletons. Twenty-three of the 37 missense variants of APOB were also found as singletons.
Hereditary breast and ovarian cancer (HBOC)
BRCA1 and BRCA2 are the major susceptibility genes for HBOC. HBOC has been thought to be less prevalent in the East Asian countries, including Japan [25]. However, reports of germline genetic variations among patients with breast and ovarian cancers indicated that the population frequency of susceptible genetic variants of HBOC may be higher than that previously thought in the Japanese population [26, 27].
In 2KJPN, we identified three nonsense variants (one in BRCA1 and two in BRCA2) that were reported as responsible variants (Table 3). A nonsense variant, BRCA1 p.Leu63*, was found in a heterozygous individual in 2KJPN [28]. Interestingly, the BRCA1 p.Arg1699Gln variant is known to be one of the genetic risk factors for intermediate breast and ovarian cancers [29], and this variant was found in three heterozygous individuals in 2KJPN (MAF = 0.07%). A missense variant, BRCA1 p.Val271Met (rs80357244), was classified as DM in HGMD and was found in 27 heterozygous individuals (MAF = 0.7%) in 2KJPN. However, this variant was categorized as variant of uncertain significance (VUS) in ClinVar, and was also classified as “polymorphic” by FALCO biosynthesis [26], a private genetic testing company. Therefore, this allele may not have a strong effect on HBOC susceptibility. This allele was found only in East Asians (in the ExAC database) and not in EAs and AAs.
Additionally, two nonsense variants in BRCA2 were reported as pathogenic and were found in 2KJPN (Table 3). BRCA2 p.Arg2318* is also one of the known mutations of HBOC in Japan [30], and a heterozygous individual was identified in 2KJPN. The other nonsense variant (p.Arg3384*) in BRCA2 was identified in two heterozygous individuals. In a previous study [31], this variant (p.Arg3384*) did not result in any cancer predisposition and was classified as a possibly benign variation, probably because this nonsense variant is located at the C-terminus end of the gene product. On the contrary, a missense variant of BRCA2, p.Ile2675Val, was detected in a heterozygous individual in 2KJPN and is considered to be a pathogenic variant affecting splicing [32]. Additionally, BRCA2 p.Gly2044Val, also registered as DM in HGMD, was found in 59 heterozygous individuals in 2KJPN (MAF = 1.4%), and has been classified as “polymorphic” by FALCO biosynthesis [26].
These results suggest that the population frequencies of susceptible variants of HBOC might be much higher in the Japanese population than previously thought, even though we have not included insertions and deletions in our analysis. Because multiple structural variants of BRCA1 and BRCA2 associated with HBOC are reported [33], checking for the presence or absence of these variations is essential. Several missense variants with high scores of pathogenicity, such as BRCA2 p.Gly2508Ser (CADD phred = 35 and M-CAP score [24] = 0.204), would require further studies to determine its association with HBOC, although most of the reported variants with strong effects in the two genes are nonsense or frame-shifting variants.
Lynch syndrome genes
Lynch syndrome (LS) is known to have a familial predisposition of colon cancer accompanied with stomach and endometrial cancers; 3% of newly diagnosed colorectal cancers develop due to LS [34]. Most of the genes responsible for LS are related to the DNA mismatch repair function and are inherited in an autosomal dominant manner [34]. Molecular diagnoses of LS would contribute to the early diagnosis of cancers in multiple organs, and are critical for the treatment of cancers in patients with LS. MSH2 and MLH1 are the major susceptible genes for LS; around 70% of the reported mutations for LS were found in these genes [35]. Most of the reported susceptible variants of LS are nonsense mutations, out-of-frame indels, or splicing error-causing variants. One important feature of the mutations found in LS-susceptible variants is that most of them are unique for each family [35].
There are six RP variants in MSH2 in 2KJPN (Table 3). An RP variant, p. Leu811*, has been reported in a few Japanese families with LS [36,37,38]. The other five variants labeled as RP, such as rs138068023 (c.-68-157G > C) and p.Pro5Gln, are not listed in either ClinVar or InSiGHT databases (see review by Chao et al. 2008 [39]); many of them are not rare in 2KJPN so that most of them would not be pathogenic.
In MLH1, there are six RP variants in 2KJPN (Table 3). An intronic variant, c.1668-1 G > A, found in a heterozygous individual in 2KJPN, may cause exon skipping and is considered to be pathogenic [40]. Another missense variant, p.Arg687Trp, found in a heterozygous individual, was previously reported in two Japanese families with LS [41]. Other RP variants are considered to be not pathogenic or of unknown significance in other databases. For example, a missense variant, p.Arg385Cys, is annotated as “likely pathogenic” in the ClinVar database and a report shows co-segregation with cancer phenotype [42]. However, the same research group later stated that the variant was a “missense variant of unreported pathogenicity” [43]. This variant was found in five individuals in 2KJPN, and further studies are needed to clarify the pathogenic roles of this variant. Similarly, p. Leu582Val is classified as “reported pathogenic” but Takahashi et al. (2007) reported that this variant, which was found in six individuals in 2KJPN, had no functional significance [44]. Another missense variant, p.Gln701Lys, was classified as an RP variant, which is a rare variant, but is considered “likely benign” by InSiGHT database [45].
Retinoblastoma
Retinoblastoma (Rb) is the most frequent intraocular malignant tumor in children, with an incidence rate ranging from 1/15,000 to 1/18,000 live births [46]. Rb is caused by bi-allelic inactivation of RB1 located on chromosome 13q14 that encodes RB protein. In the dominantly inherited form, one mutation is inherited through the germline and the secondary mutation occurs in the somatic cells [47]. The Rb protein acts as a tumor suppressor, which regulates cell growth and stops cells from undergoing uncontrolled proliferation.
In 2KJPN, we found two missense variants of RB1, p.Arg621Ser (rs367578442, CADD phred score = 12.5) and p.Leu819Val (CADD phred score = 15.26) as canP variants, which are registered in HGMD in the “DM?” category (Table 3), and were originally reported by a previous study on Chinese patients with Rb [48]. The p.Arg621Ser variant was found in two heterozygous individuals, and p.Leu819Val was found in a heterozygous individual.
Furthermore, p.Arg621Ser is located between two RB1 pocket-domains, which feature an additional protein-binding site [49], and p.Leu819Val is located in the RB1 C-terminus region, which is involved in association with the dimer surface resulting from an association of the E2 factors (E2Fs) [50]. The p.Arg621Ser variant was also found in an AA subject in an ancestrally diverse cohort of 681 healthy individuals [51]. A previous study involving Japanese patients with Rb found that a majority of the somatic mutations were found in the adenovirus early region 1A (ElA) binding sites [52]; however, the p.Arg621Ser and p.Leu819Val variants have not been reported in any domestic report in patients with Rb. Further examination of genetic variants in the germline may be needed to assess the frequency of the risk variants in RB1 in the Japanese population. In addition to inactivation of Rb1 itself, deregulation of Rb1-related biological pathways has critical roles in most types of human cancer. A more precise annotation and identification of RB1 mutations could play a pivotal role in enhancing the clinical management of the risks for Rb.
Multiple endocrine neoplasia type 2 and familial medullary thyroid cancer (FMTC)
RET is an important gene related to several clinically distinct diseases. Gain-of-function mutations in RET cause multiple endocrine neoplasia (MEN) type 2 and FMTC [53]. On the contrary, loss-of-function mutations in RET are known to be risk factors for Hirschsprung disease (HSCR), which is caused by the congenital absence of parasympathetic ganglion cells in the intestinal tissues [54]. Furthermore, several mutations of this gene have also been reported in patients with congenital central hypoventilation syndrome (CCHS) [55]. In 2KJPN, known missense variants, p.Val292Met, for MEN type 2 (MAF = 0.0015), and p.Gly321Arg, for FMTC (MAF = 0.00049), were found as variants causing genetically dominant effects (Table 3). Our results suggest that screening the sequence of RET may be beneficial for early recognition of patients with MEN type 2 and FMTC in the Japanese population.
In addition, we found one missense variant, p.Arg114His, classified as DM in HGMD for CCHS as variants with loss-of-function effects. The allele frequency of this variant (MAF = 0.0056) in 2KJPN was higher than that of EAs (p < 0.00001) (see Supplementary Table 3 for inter-ethnic comparisons), and this variant was originally reported in a domestic study [56]. In ClinVar, this variant was annotated with “conflicting interpretations of pathogenicity.” Further studies are needed to clarify the pathogenic role of this variant. We also found another missense variant, p.Thr278Asn, for HSCR (MAF = 0.012) that was registered as DM in HGMD. However, this variant was annotated with “conflicting interpretations of pathogenicity in the ClinVar database.” This variant was originally reported in Asia [56], and the allele frequency in people with European and African ancestries is very low (not detected in ESP EA and AA). Further studies are needed to clarify the clinical impact of this variant.
Familial hypercholesterolemia
Familial hypercholesterolemia (FH) is a relatively common genetic disorder with a prevalence of 1:200–500. Generally, people with untreated FH are at a higher risk of coronary heart disease [57]. Genetic variants in three genes—low-density lipoprotein receptor (LDLR) [58], APOB, and proprotein convertase subtilisin/kexin type 9 (PCSK9) [59]—account for the majority of cases with autosomal dominant FH [60].
LDLR
The LDLR protein recognizes apolipoprotein B-100 (apo B-100) embedded in the outer phospholipid layer of low-density lipoproteins (LDLs), and mediates the endocytosis of LDL. After internalization of the LDLR-LDL complexes into the endosomes, the complexes dissociate and LDLR is either recycled or degraded, whereas LDL is taken into lysosomes where the protein moiety is degraded. LDLR variants that cause FH result in defective synthesis, assembly, LDL-binding, transport, or recycling of the protein, causing reduced clearance of LDL, the major plasma cholesterol-carrier, and thus, dramatically raising blood cholesterol levels.
In search for potentially pathological variants of FH in the 2KJPN reference panel, we identified four missense SNVs (p.Arg115His, p.Arg257Trp, p.Leu568Val, and p.Pro699Leu) classified as DM in HGMD (Table 3). Three of these variants, p.Arg115His, p.Leu568Val, and p.Pro699Leu, are classified as “likely pathogenic” in ClinVar. An additional missense SNV, p.Gly461Ser, was classified as “likely pathogenic” in ClinVar and was identified in 2KJPN, but no annotations were given to this variant in HGMD. Through a literature survey (Table 4), three of these variants (p.Arg257Trp, p.Leu568Val, and p.Pro699Leu found in two, one, and one heterozygous individuals, respectively) identified in 2KJPN were found to have strong evidence for the pathological significance for FH. This finding was based on multiple studies with patients of European or East Asian origin, including the Japanese, and significantly higher allele frequencies in these patients were identified over the population controls, including those in 2KJPN. In regards to another missense variant, p.Gly461Ser (found in two heterozygous individuals in 2KJPN), only one report of two probands with this variant in 262 Greek families with FH was found [61], suggesting a higher allele frequency of this variant in patients over the general population, as this variant was not found in over 70,000 Europeans in the Exome Aggregation Consortium (ExAC) [62]. However, the pathological significance of this variant should be confirmed by further studies. As for another missense variant, p.Arg115His, the allele frequency in 2KJPN was 0.0039, which was greater than expected for causative variants of FH based on the estimated prevalence of 1 in 200–500 in Japan, hence suggesting that these variants are benign or have mild effects. These two variants showed a higher allele frequency in 2KJPN than that of EAs (p < 0.00001), and both of them were originally reported from domestic studies [63, 64]. Thus, these variants could be classified as benign evaluated solely from the viewpoint of their relatively high frequencies.
APOB
APOB mutations have been estimated to account for 1–5% of patients with FH, and are inherited in an autosomal dominant manner [65, 66]. From APOB, apo B-100 is synthesized exclusively in the liver as one of the two main protein isoforms, and is a major constituent of LDL and VLDL. Apo B-100 serves as a recognition signal for the LDL receptor to bind and internalize LDL particles. Furthermore, APOB pathogenic variants decrease the binding affinity of LDL particles for the LDL receptor, thus causing fewer LDL particles to be cleared from the blood, which then dramatically raises the plasma cholesterol levels.
In the 2KJPN reference panel, we identified two reported pathogenic missense variants (Table 3). One of the variants, p.Arg3527Gln, is a well-characterized pathogenic missense variant [67] in APOB, and was identified in only one heterozygous individual among the 2049 individuals (allele frequency = 0.0002). Another missense variant, p.Ile3768Thr [68], was registered as DM in HGMD, and was identified in 2KJPN in a heterozygous individual. However, no evidence for the pathogenicity of this variant has been presented; therefore, its clinical and functional significance must be scrutinized.
PCSK9
PCSK9 encodes neural apoptosis regulated convertase (NARC)-1, a 692 AA protein that is the ninth member of the secretory subtilase family [69]. The protein is synthesized as a soluble zymogen that undergoes autocatalytic cleavage in the endoplasmic reticulum. The mature protein binds to the EGF-A domain of lipoprotein receptors [70, 71], abolishes their functions, and raises the level of cholesterol in the blood stream. Furthermore, some gain-of-function mutations increase the binding affinity of PCSK9 and lipoprotein receptors, thus resulting in the degradation of LDLR, inefficient incorporation of cholesterol in the liver cells, and higher cholesterol levels in the blood stream. On the contrary, individuals having loss-of-function variants showed lower levels of LDL cholesterol [72].
In the 2KJPN reference panel, we identified six reported pathogenic SNVs for PCSK9—five missense variants (p.Glu54Ala, p.Arg104Cys, p.Arg215His, p.Ala514Thr, and p.Ser668Arg) and one nonsense variant (p.Trp428*) (Table 3). Since p.Ser668Arg and p.Trp428* were reported to be causative variants for low LDL cholesterol [63], we considered the other four missense variants (p.Glu54Ala, p.Arg104Cys, p.Arg215His, and p.Ala514Thr) as causative variants for hypercholesterolemia. The population frequency of the causative variants responsible for hypercholesterolemia in PCSK9 was estimated to be 0.001 in 2KJPN, which was based on the allele frequencies of the four responsible variants, all of which were singletons. There was a missense variant, p.Glu32Lys (rs564427867), which was registered as DM in HGMD, and this variant was found in 2KJPN in 44 heterozygous and one homozygous individual. Although this variant was discarded during variant filtering due to its relatively higher frequency (0.011) for assigning RP variants, we further reviewed this variant because it was reported in a domestic study [73]. In ClinVar, this variant was annotated as “conflicting interpretations of pathogenicity.” Because it showed a higher allele frequency in 2KJPN than that in non-Asian populations (p < 0.00001), the variant might not have been detected or reported by DNA sequencing of the patient samples from non-Asian population (see Supplementary Table 3). The variant could be classified as benign based on its high frequency; however, it may have mild effects on FH. Our results showed that phenotypic variants responsible for high and low levels of LDL cholesterol were found in 2KJPN. Further careful examination may be needed to assess the proportion of risk variants in LDLR, APOB, and PCSK9 in the Japanese population [74]. It is very important to know which genes have causative mutations in patients with FH to assess for appropriate therapeutic strategies [75] for personalized medicine.
Discussion
Here, we presented the estimation of pathogenic variant frequencies for actionable genes in the Japanese population for the first time, and showed that a substantial number of people had reported pathogenic variants of the ACMG-recommended genes. Although there have been numerous domestic reports on pathogenic variants of diseases detected in patient groups, it was not clear in what proportion the responsible variants exist among healthy individuals. Identification of potential risk alleles and their frequency estimation among healthy individuals are, thus, highly important for public health.
We also found that manually checking and reviewing variants are very critical to interpreting variants for its pathogenicity, although it is needless to say about distinguishing distinct phenotypic effects by single genes, such as RET and PCSK9. In this study, for several diseases, we manually reviewed pathogenic variants annotated in public databases (HGMD and ClinVar). We conducted a careful review of the variants, especially for the three genes (LDLR, APOB, PCSK9) responsible for FH, and the allelic frequencies of the risk alleles were compared with the prevalence data in the Japanese population for each condition. We found that evidence of pathogenicity in the Japanese population was lacking for some variants, even if they were reported as DMs, such as in the case of genes responsible for FH. In addition, we observed that some of the reported pathogenic variants could be benign after a review of the variants. The insufficiencies in the data of reported pathogenic variants may be due to an insufficient examination of allele frequency in healthy controls in the original studies or an inappropriate curation in the public databases.
We found that some of HGMD-DM variants existed at higher frequencies in 2KJPN than in other ethnic groups. The examples were RET (p.Arg114His and p.Thr278Asn), LDLR (p.Arg115His), and PCSK9 (p.Glu32Lys) among the genes in our variant review and inter-ethnic allele frequency comparison. These examples may suggest that protein-altering variants, such as missense or truncating variants, which exist in Asian populations, but are rarely detected in other ethnic populations, are more likely to be reported in the literature as novel DMs for Asians. Although these variants may have mild effects on phenotypes, we should re-review reported pathogenic variants to check whether protein-altering SNVs showing inter-ethnic frequency differences have been biasedly reported or registered in the databases as novel pathological variants.
Although public databases of pathogenic variants, such as HGMD and ClinVar, are useful as information resources, reviewing reported pathogenic variants for their pathogenicity in the target population is necessary and a challenging issue. In particular, this is critical for returning individual genomic results with clinical benefits and avoiding unnecessary psychosocial harm due to uncertain clinical validity. Our study suggested that some of the reported pathogenic variants should be re-reviewed, even though they were designated as “disease-causing variants”, especially when they are used for the purposes of identification of secondary findings in clinical settings under the ACMG recommendations.
Several previous reports have tried to overcome this issue, and one of these studies estimated the frequency of actionable variants in the diverse 1000 genomes [15]. They conducted an extensive literature survey by checking the population frequency, evidence for pathogenicity, and evaluations by expert physicians with medical specialties relevant to the conditions. Although 237 variants were annotated as disease-causing variants by HGMD, only seven variants remained to be likely pathogenic after the variant review.
Information of individual status of risk variants should be utilized for public health. The participants in our cohort studies were very interested in individual genomic results [76]. However, in the present situation in Japan, actual attempts or trials of returning individual genomic results in the research context have been very limited. This may be due to a number of medical, psychosocial, ethical, and financial issues, as well as the lack of experiences. Such situations may vary among countries, and real actionability depends on the medical systems in the society. Considering the current situation in Japan, we have a plan of action to return individual genomic results to the participants of our cohort studies (will be described elsewhere). We may be able to follow their medical conditions in the long-term after the participants receive their genomic results [77]. We expect that this kind of practice would contribute to the accumulation of case information about dealing with genetic results from the standpoints of scientific and practical aspects.
Furthermore, there are limitations in this study because 1) insertions and deletions were not analyzed; 2) actionable genes in chromosome X (GLA and OTC) were not analyzed; and 3) reported pathogenic variants were not detected for 15 genes, which may be due to the limited number of individuals or very low allele frequencies in the Japanese population. Although we obtained variants as candidates of expected pathogenic variants, further analysis and evaluation through appropriate filtering and interpretations are needed for selecting strong candidates of pathogenic variants. We would overcome these limitations as much as possible by including other types of variants and extending our analysis in the near future.
It may be not surprising that about 21% people have reported pathogenic variants of the 57 ACMG genes. In our previous study with 1KJPN, we showed that one individual had 11.2 HGMD-DM variants (9.6 as heterozygous and 1.6 as homozygous) on average [18]. In this study, a small fraction of the low-frequency HGMD-DM variants in the limited set of disease genes may have been detected. Such estimates may be lowered if variants were reviewed appropriately for all the target genes.
The recommended gene list for incidental findings, which were originally proposed by ACMG, may be modified by considering its practicality for East Asian populations. For example, a Korean group is considering inclusion of CDH1 for the risk of hereditary diffuse gastric cancer [11], based on its high penetrance. This kind of consideration would improve the quality of genetic medicine in East Asian countries. Other phenotypes not listed in the recommendations by ACMG may be taken into consideration if they are important for healthcare in the Japanese population. Based on this study, we should construct an information infrastructure of pathogenic variants for the Japanese population. Through appropriate variant interpretations, updated information of pathogenic variants would be useful for diagnostic strategies and subsequent personalized healthcare for the Japanese population.
References
Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46:818–25.
Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47:435–44.
UK 10K Consortium, Walter K, Min JL, Huang J, Crooks L, Memari Y, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90.
Styrkarsdottir U, Thorleifsson G, Sulem P, Gudbjartsson DF, Sigurdsson A, Jonasdottir A, et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature. 2013;497:517–20.
Koshiba S, Motoike I, Kojima K, Hasegawa T, Shirota M, Saito T, et al. The structural origin of metabolic quantitative diversity. Sci Rep. 2016;6:31463.
Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9.
Higasa K, Miyake N, Yoshimura J, Okamura K, Niihori T, Saitsu H, et al. Human genetic variation database, a reference database of genetic variations in the Japanese population. J Hum Genet. 2016;61:547–53.
Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74.
Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2016;19:249–55.
Jurgens J, Ling H, Hetrick K, Pugh E, Schiettecatte F, Doheny K, et al. Assessment of incidental findings in 232 whole-exome sequences from the Baylor-Hopkins Center for Mendelian Genomics. Genet Med. 2015;17:782–88.
Jang MA, Lee SH, Kim N, Ki CS. Frequency and spectrum of actionable pathogenic secondary findings in 196 Korean exomes. Genet Med. 2015;17:1007–11.
Gambin T, Jhangiani SN, Below JE, Campbell IM, Wiszniewski W, Muzny DM, et al. Secondary findings and carrier test frequencies in a large multiethnic sample. Genome Med. 2015;7:54.
Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–79.
Dorschner MO, Amendola LM, Turner EH, Robertson PD, Shirts BH, Gallego CJ, et al. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am J Hum Genet. 2013;93:631–40.
Olfson E, Cottrell CE, Davidson NO, Gurnett CA, Heusel JW, Stitziel NO, et al. Identification of Medically Actionable Secondary Findings in the 1000 Genomes. PloS ONE. 2015;10:e0135193.
Tabor HK, Auer PL, Jamal SM, Chong JX, Yu JH, Gordon AS, et al. Pathogenic variants for Mendelian and complex traits in exomes of 6,517 European and African Americans: implications for the return of incidental results. Am J Hum Genet. 2014;95:183–93.
Kuriyama S, Yaegashi N, Nagami F, Arai T, Kawaguchi Y, Osumi N, et al. The Tohoku Medical Megabank project: design and mission. J Epidemiol. 2016;26:493–511.
Nagasaki M, Yasuda J, Katsuoka F, Nariai N, Kojima K, Kawai Y, et al. Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat Commun. 2015;6:8018.
Yamaguchi-Kabata Y, Nariai N, Kawai Y, Sato Y, Kojima K, Tateno M, et al. iJGVD: an integrative Japanese genome variation database based on whole-genome sequencing. Hum Genome Var. 2015;2:15050.
Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–81.
Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–68.
Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–15.
Jagadeesh KA, Wenger AM, Berger MJ, Guturu H, Stenson PD, Cooper DN, et al. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet. 2016;48:1581–86.
Liede A, Narod SA. Hereditary breast and ovarian cancer in Asia: genetic epidemiology of BRCA1 and BRCA2. Hum Mutat. 2002;20:413–24.
Hirotsu Y, Nakagomi H, Sakamoto I, Amemiya K, Mochizuki H, Omata M. Detection of BRCA1 and BRCA2 germline mutations in Japanese population using next-generation sequencing. Mol Genet Genomic Med. 2015;3:121–9.
Sakamoto I, Hirotsu Y, Nakagomi H, Ouchi H, Ikegami A, Teramoto K, et al. BRCA1 and BRCA2 mutations in Japanese patients with ovarian, fallopian tube, and primary peritoneal cancer. Cancer. 2016;122:84–90.
Sekine M, Nagata H, Tsuji S, Hirai Y, Fujimoto S, Hatae M, et al. Mutational analysis of BRCA1 and BRCA2 and clinicopathologic analysis of ovarian cancer in 82 ovarian cancer families: two common founder mutations of BRCA1 in Japanese population. Clin Cancer Res. 2001;7:3144–50.
Spurdle AB, Whiley PJ, Thompson B, Feng B, Healey S, Brown MA, et al. BRCA1 R1699Q variant displaying ambiguous functional abrogation confers intermediate breast and ovarian cancer risk. J Med Genet. 2012;49:525–32.
Ikeda N, Miyoshi Y, Yoneda K, Shiba E, Sekihara Y, Kinoshita M, et al. Frequency of BRCA1 and BRCA2 germline mutations in Japanese breast cancer families. Int J Cancer. 2001;91:83–8.
Borg A, Haile RW, Malone KE, Capanu M, Diep A, Torngren T, et al. Characterization of BRCA1 and BRCA2 deleterious mutations and variants of unknown clinical significance in unilateral and bilateral breast cancer: the WECARE study. Hum Mutat. 2010;31:E1200–40.
Bonnet C, Krieger S, Vezain M, Rousselin A, Tournier I, Martins A, et al. Screening BRCA1 and BRCA2 unclassified variants for splicing mutations using reverse transcription PCR on patient RNA and an ex vivo assay based on a splicing reporter minigene. J Med Genet. 2008;45:438–46.
Walsh T, Lee MK, Casadei S, Thornton AM, Stray SM, Pennil C, et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci USA. 2010;107:12629–33.
Tiwari AK, Roy HK, Lynch HT. Lynch syndrome in the 21st century: clinical perspectives. QJM. 2016;109:151–8.
Peltomaki P. Update on Lynch syndrome genomics. Fam Cancer. 2016;15:385–93.
Lu SL, Akiyama Y, Nagasaki H, Nomizu T, Ikeda E, Baba S, et al. Loss or somatic mutations of hMSH2 occur in hereditary nonpolyposis colorectal cancers with hMSH2 germline mutations. Jpn J Cancer Res. 1996;87:279–87.
Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Onda A, Okumura Y, et al. Molecular nature of colon tumors in hereditary nonpolyposis colon cancer, familial polyposis, and sporadic colon cancer. Gastroenterology. 1996;111:307–17.
Miyaki M, Konishi M, Muraoka M, Kikuchi-Yanoshita R, Tanaka K, Iwama T, et al. Germ line mutations of hMSH2 and hMLH1 genes in Japanese families with hereditary nonpolyposis colorectal cancer (HNPCC): usefulness of DNA analysis for screening and diagnosis of HNPCC patients. J Mol Med. 1995;73:515–20.
Chao EC, Velasquez JL, Witherspoon MS, Rozek LS, Peel D, Ng P, et al. Accurate classification of MLH1/MSH2 missense variants with multivariate analysis of protein polymorphisms-mismatch repair (MAPP-MMR). Hum Mutat. 2008;29:852–60.
Arnold S, Buchanan DD, Barker M, Jaskowski L, Walsh MD, Birney G, et al. Classifying MLH1 and MSH2 variants using bioinformatic prediction, splicing assays, segregation, and tumor characteristics. Hum Mutat. 2009;30:757–70.
Furukawa T, Konishi F, Shitoh K, Kojima M, Nagai H, Tsukamoto T. Evaluation of screening strategy for detecting hereditary nonpolyposis colorectal carcinoma. Cancer. 2002;94:911–20.
Cravo M, Afonso AJ, Lage P, Albuquerque C, Maia L, Lacerda C, et al. Pathogenicity of missense and splice site mutations in hMSH2 and hMLH1 mismatch repair genes: implications for genetic testing. Gut. 2002;50:405–12.
Lage PA, Albuquerque C, Sousa RG, Cravo ML, Salazar M, Francisco I, et al. Association of colonic and endometrial carcinomas in Portuguese families with hereditary nonpolyposis colorectal carcinoma significantly increases the probability of detecting a pathogenic mutation in mismatch repair genes, primarily the MSH2 gene. Cancer. 2004;101:172–7.
Takahashi M, Shimodaira H, Andreutti-Zaugg C, Iggo R, Kolodner RD, Ishioka C. Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res. 2007;67:4595–604.
Thompson BA, Spurdle AB, Plazzer JP, Greenblatt MS, Akagi K, Al-Mulla F, et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet. 2014;46:107–15.
Bishop JO, Madson EC. Retinoblastoma. Review of the current status. Surv Ophthalmol. 1975;19:342–66.
Knudson AG Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–3.
He MY, An Y, Gao YJ, Qian XW, Li G, Qian J. Screening of RB1 gene mutations in Chinese patients with retinoblastoma and preliminary exploration of genotype-phenotype correlations. Mol Vis. 2014;20:545–52.
Lee JO, Russo AA, Pavletich NP. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–65.
Rubin SM, Gall AL, Zheng N, Pavletich NP. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell. 2005;123:1093–106.
Bodian DL, McCutcheon JN, Kothiyal P, Huddleston KC, Iyer RK, Vockley JG, et al. Germline variation in cancer-susceptibility genes in a healthy, ancestrally diverse cohort: implications for individual genome sequencing. PloS ONE. 2014;9:e94554.
Shimizu T, Toguchida J, Kato MV, Kaneko A, Ishizaki K, Sasaki MS. Detection of mutations of the RB1 gene in retinoblastoma patients by using exon-by-exon PCR-SSCP analysis. Am J Hum Genet. 1994;54:793–800.
Krampitz GW, Norton JA. RET gene mutations (genotype and phenotype) of multiple endocrine neoplasia type 2 and familial medullary thyroid carcinoma. Cancer. 2014;120:1920–31.
Pan ZW, Li JC. Advances in molecular genetics of Hirschsprung’s disease. Anat Rec. 2012;295:1628–38.
Sasaki A, Kanai M, Kijima K, Akaba K, Hashimoto M, Hasegawa H, et al. Molecular analysis of congenital central hypoventilation syndrome. Hum Genet. 2003;114:22–6.
Kanai M, Numakura C, Sasaki A, Shirahata E, Akaba K, Hashimoto M, et al. Congenital central hypoventilation syndrome: a novel mutation of the RET gene in an isolated case. Tohoku J Exp Med. 2002;196:241–6.
Emerging Risk Factors, C, Di Angelantonio E, Sarwar N, Perry P, Kaptoge S, Ray KK, et al. Major lipids, apolipoproteins, and risk of vascular disease. JAMA. 2009;302:1993–2000.
Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47.
Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–6.
Henderson R, O’Kane M, McGilligan V, Watterson S. The genetics and screening of familial hypercholesterolaemia. J Biomed Sci. 2016;23:39.
Mollaki V, Progias P, Drogari E. Familial Hypercholesterolemia in Greek children and their families: genotype-to-phenotype correlations and a reconsideration of LDLR mutation spectrum. Atherosclerosis. 2014;237:798–804.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Miyake Y, Yamamura T, Sakai N, Miyata T, Kokubo Y, Yamamoto A. Update of Japanese common LDLR gene mutations and their phenotypes: Mild type mutation L547V might predominate in the Japanese population. Atherosclerosis. 2009;203:153–60.
Varret M, Rabes JP, Thiart R, Kotze MJ, Baron H, Cenarro A, et al. LDLR Database (second edition): new additions to the database and the software, and results of the first molecular analysis. Nucleic Acids Res. 1998;26:248–52.
Soria LF, Ludwig EH, Clarke HR, Vega GL, Grundy SM, McCarthy BJ. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci USA. 1989;86:587–91.
Vega GL, Grundy SM. In vivo evidence for reduced binding of low density lipoproteins to receptors as a cause of primary moderate hypercholesterolemia. J Clin Invest. 1986;78:1410–14.
Braenne I, Kleinecke M, Reiz B, Graf E, Strom T, Wieland T, et al. Systematic analysis of variants related to familial hypercholesterolemia in families with premature myocardial infarction. Eur J Hum Genet. 2016;24:191–7.
Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–7.
Seidah NG, Benjannet S, Wickham L, Marcinkiewicz J, Jasmin SB, Stifani S, et al. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc Natl Acad Sci USA. 2003;100:928–33.
Kwon HJ, Lagace TA, McNutt MC, Horton JD, Deisenhofer J. Molecular basis for LDL receptor recognition by PCSK9. Proc Natl Acad Sci USA. 2008;105:1820–5.
Cunningham D, Danley DE, Geoghegan KF, Griffor MC, Hawkins JL, Subashi TA, et al. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nat Struct Mol Biol. 2007;14:413–9.
Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–72.
Noguchi T, Katsuda S, Kawashiri MA, Tada H, Nohara A, Inazu A, et al. The E32K variant of PCSK9 exacerbates the phenotype of familial hypercholesterolaemia by increasing PCSK9 function and concentration in the circulation. Atherosclerosis. 2010;210:166–72.
Abul-Husn NS, Manickam K, Jones LK, Wright EA, Hartzel DN, Gonzaga-Jauregui C, et al. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science. 2016;354:aaf7000.
Fitzgerald K, Frank-Kamenetsky M, Shulga-Morskaya S, Liebow A, Bettencourt BR, Sutherland JE, et al. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet. 2014;383:60–8.
Ishikuro M, Nakaya N, Obara T, Sato Y, Metoki H, Kikuya M, et al. Public attitudes toward an epidemiological study with genomic analysis in the Great East Japan Earthquake disaster area. Prehosp Disaster Med. 2016;31:330–4.
Wald DS, Bestwick JP, Morris JK, Whyte K, Jenkins L, Wald NJ. Child-parent familial hypercholesterolemia screening in primary care. N Engl J Med. 2016;375:1628–37.
Acknowledgements
This work was supported by Tohoku Medical Megabank Project from MEXT and Japan Agency for Medical Research and Development (AMED), and by the grant “Advanced Genome Research and Bioinformatics Study to Facilitate Medical Innovation”, also by AMED. This research was also supported by the Center of Innovation Program from Japan Science and Technology Agency, JST. All computational resources were provided by the ToMMo supercomputer system. We are indebted to all volunteers who participated in this Tohoku Medical Megabank project. We would like to acknowledge all the members associated with this project; the member list is available at the following web site: http://www.megabank.tohoku.ac.jp/english/a161201/. We would like to thank Editage (www.editage.jp) for English language editing.
Author information
Authors and Affiliations
Consortia
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Yamaguchi-Kabata, Y., Yasuda, J., Tanabe, O. et al. Evaluation of reported pathogenic variants and their frequencies in a Japanese population based on a whole-genome reference panel of 2049 individuals. J Hum Genet 63, 213–230 (2018). https://doi.org/10.1038/s10038-017-0347-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-017-0347-1
This article is cited by
-
Assessing the efficacy of target adaptive sampling long-read sequencing through hereditary cancer patient genomes
npj Genomic Medicine (2024)
-
Assessment of pathogenic variation in gynecologic cancer genes in a national cohort
Scientific Reports (2023)
-
Frequency of actionable secondary findings in 7472 Korean genomes derived from the National Project of Bio Big Data pilot study
Human Genetics (2023)
-
Comparison of the loci associated with HbA1c and blood glucose levels identified by a genome-wide association study in the Japanese population
Diabetology International (2023)
-
Pathological variants in genes associated with disorders of sex development and central causes of hypogonadism in a whole-genome reference panel of 8380 Japanese individuals
Human Genome Variation (2022)