Abstract
Insulin, IGF, and relaxin are established members of the insulin protein superfamily. The application of the techniques of cellular, molecular, and computational biology has permitted the identification of new insulin-like ligands and their cognate receptors. Information regarding the biologic role is available for some of these newly identified ligand-receptor systems and indicates novel roles in diverse processes such as testicular descent, germ cell function, and cell migration.
Similar content being viewed by others
Main
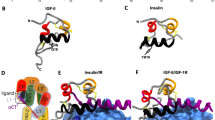
The insulin/IGF/relaxin superfamily is an ancient family of functionally diverse proteins. Insulin or insulin-like proteins have been described in unicellular eukaryotes as well as in such primitive species as insects, tunicates, annelids, and molluscans (1–6). Despite the broad magnitude of functional divergence present within the family, all proteins of the insulin family exhibit a high degree of structural conservation (Fig. 1). The primary peptide sequence of each member of the family is characterized by three domains comprised of an amino terminal B peptide (or chain) joined to a carboxyl A peptide by an intervening C peptide (B-C-A) (7). Between the different hormones within the family (paralog) and similar hormones in different species (ortholog), the B and A chain peptides are relatively invariant and exhibit a pattern of distinct and highly conserved cysteine motifs. These cysteine motifs characterize the family; specifically the motif (CC-3X-C-8X-C) present in the A peptide has been termed the insulin signature. Many members of the insulin family of hormones are synthesized as preprohormones, with the primary peptide undergoing posttranslational modification to generate a cysteine-linked heterodimer of the B and A peptides that functions as the active hormone.
A decade ago the insulin family was comprised of four members in mammals: insulin, IGF-I, IGF-II, and relaxin. In recent years, additional members of the family, termed INSL3 (8,9), INSL4 (10,11), INSL5 (12,13), INSL6 (13–15), and INSL7 (16), have been identified (17) (Fig. 2). The discovery of these new ligands spurred efforts to identify their cognate receptors. These efforts resulted in the discovery that LGR8, a member of the LGR (leucine-rich repeat-containing G protein-coupled receptors) subfamily of GPCR (G protein-coupled receptors) receptors, was the cognate receptor for INSL3 (18). This review will describe new information regarding these new members of the insulin family and their cognate receptors with emphasis on the putative biologic role(s) of these proteins.
INSL3
Having been initially identified in boar testes (19), the product of the INSL3 gene, relaxin-like factor, or Ley-I-L (Leydig insulin-like peptide precursor), was subsequently identified in both the human and the mouse as a single copy gene containing two exons and a single intron (8,20). Human INSL3 is located on the short arm of chromosome 19 and is structurally similar to relaxin. The major site of expression of INSL3 is in Leydig cells of the testes, but it is also expressed in the theca cells of the corpus luteum, the trophoblast, breast, and a variety of other tissues (21–25). The first indication of an essential role for INSL3 in reproduction came from studies of knockout models in mice. Insl3 null male mice were cryptorchid suggesting that Insl3 gene plays a major role in the development of the gubernaculum and subsequent testicular development (9,26). The phenotype of the Insl3 null mice was similar to that of the LGR8 (GREAT) null mice, leading to identification of LGR8 as the cognate receptor for INSL3 (18,27). Subsequent screening of boys with cryptorchidism, the most frequent congenital abnormality in humans, resulted in the identification of mutations within the INSL3 gene and its promoter sequence and the LGR8 gene (28–30), although the mutations in these two genes are responsible for only a small proportion (<10%) of familial cryptorchidism (31). The contribution of abnormalities in LGR expression, cognate signaling pathways, and target genes in the pathogenesis of cryptorchidism remains to be investigated.
In the testis, in addition to its actions on the gubernaculum, INSL3 suppresses germ cell apoptosis and acts in a paracrine manner as a survival factor for male germ cells (32). Whereas much of the interest regarding the biologic role of INSL3 has been focused on the testes, recent experimental observations provide clues to actions of INSL3 in the ovary. LH transiently increases INSL3 expression in theca cells of the ovary and INSL3 suppresses intra-oocyte cAMP levels with stimulation of induction of oocyte maturation, suggesting a paracrine role for INSL3 in mediating preovulatory LH actions on the ovary (32).
INSL4
Efforts directed at identifying differentially expressed genes within the human placenta led to the identification of the product of the INSL4 gene, early placenta insulin-like peptide (EPIL) or placentin (10,11). In contrast to other members of the insulin superfamily, INSL4 is primate-specific as INSL4 nucleic acid sequences are absent in rodent, horse, and lemur genomes (33). Structurally, the gene is similar to insulin, relaxin, and INSL3 and contains two exons and a single intron. It is located on 9p24, where it is clustered with the two human relaxin genes (34). The predominant site of expression of the INSL4 gene is in the placenta during early pregnancy, but it is also expressed to a lesser degree in interbone ligaments, perichondrium, and the uterus (35). Given that it is detectable in maternal serum during pregnancy, INSL4 presumably functions as a hormone. It is also likely that INSL4 has paracrine and autocrine actions. There is a paucity of information regarding the cognate receptor(s) for INSL4, although conditioned media from cells overexpressing INSL4 increases total cellular tyrosine phosphorylation via a pathway distinct from that of the insulin receptor (11). Recent reports of INSL4 being overexpressed and secreted in erb-3–positive breast cancer cells with high invasive potential and in hydatiform moles suggest a role for INSL4 in tissue invasiveness and cell migration (36).
INSL5 and INSL6
Three laboratories, working independently, identified through the application of the techniques of computational biology, two new genes: INSL5 (relaxin/insulin-like factor 2, RIF2) and INSL6 (relaxin/insulin-like factor 1, RIF1) (12–15). Human, mouse, and rat orthologs of these genes have also been identified. Both genes encode proteins that are clearly members of the insulin superfamily; they contain signal peptides and exhibit the requisite cysteine motifs. The human INSL5 gene is located on chromosome 1 and the orthologous mouse Insl5 is located on mouse chromosome 4. Human INSL6 is located in the region 9p24 in proximity to the human relaxin genes as well as to INSL 4. The mouse ortholog, Insl6, is located on chromosome 19, the chromosome that also contains the single mouse relaxin gene. In humans, INSL5 is maximally expressed in the uterus and the digestive tract, with highest levels of expression in the rectum. In the mouse, expression of mouse Insl5 is described in thymus, kidney, heart, brain, and testis. INSL6 is maximally expressed in germ cells of the testis, with lower levels of expression detectable in a variety of other tissues including intestine, thymus, kidney, uterus, ovary, spleen, breast, lung, and liver (15,37). The INSL6 peptide undergoes posttranslational modifications including glycosylation and ubiquitination (37). A paracrine role for INSL6 in modulating Sertoli cell function is suggested by studies that demonstrate augmentation of FSH-stimulated cAMP in Sertoli cells (Lu C, Menon RK, unpublished data).
LGR FAMILY OF RECEPTORS
The discovery of new putative ligands in the insulin family spurred efforts to identify their cognate receptors. These efforts resulted in the deorphanizing of a group of GPCR receptors termed LGR (38), with the discovery that LGR8 was the cognate receptor for INSL3 (18). This discovery was soon followed by the identification of LGR7 and LGR8 as cognate receptors for relaxin (39), a discovery of some significance inasmuch as the identity of the relaxin receptor(s) had eluded identification since the time relaxin was the first discovered in 1926 (40). LGR are structurally similar to receptors for gonadotrophins (LH and FSH) and TSH. These receptors are characterized by a large N-terminal extracellular domain containing leucine-rich repeat preceding a GPCR seven transmembrane spanning domain. LGR can be divided into three subgroups, with the first group consisting of the gonadotropin and TSH receptors, the second subgroup consisting of LGR4, LGR5, and LGR6, and the third subgroup constituted by LGR7 and LGR8 (41) (Fig. 3). At present, LGR4, LGR5, and LGR6 remain as orphan receptors. LGR7 and LGR8 have been identified as receptors for relaxin (39). Relaxin-3 is an additional ligand for LGR7 (42) and INSL3 a ligand for LGR8 (18). The overlapping of ligand specificity between the LGR receptors suggest that tissue-specific expression profile of these receptors may enable these ligands to have differential biologic actions in various tissues. It is of interest that the LGR4 null mice exhibits intrauterine growth retardation and embryonic and perinatal lethality (43) and the LGR5 null mice displays a phenotype similar to ankyloglossia (44), suggesting that the LGR4 and LGR5 systems may have biologic roles distinct from LGR7 and 8 (Figs 1–3). Table 1
CONCLUSIONS
In relatively rapid succession, through the application of the techniques of molecular and computational biology, five new members of the insulin family and some of their cognate receptors have been identified. The two major themes that emerge from these discoveries are the expanding role of the insulin family of proteins in reproductive physiology and the identification of the GPCR receptors as cognate receptors for some of these new ligands. It is noteworthy that a recent study also implicated the “established” members of the insulin family in male sexual differentiation (45). Thus, male mice with triple knockout of the insulin, IGF-I, and insulin-related receptor displayed ovaries and a female phenotype. The identification of GPCR receptors as cognate receptors for the newer members of the insulin family is especially noteworthy because canonical insulin/IGF-I receptors are tyrosine kinase receptors and the identification of GPCR as cognate receptors for this family of ligands represents a paradigm shift. The challenge now lies in identifying the biologic function of these ligands and receptor systems. A more complete understanding of the biologic roles of these new proteins mayenable the design of novel therapeutic strategies in the treatment of conditions such as premature labor, infertility, and malignancy.
Abbreviations
- GPCR:
-
G protein-coupled receptor
- GREAT:
-
G protein-coupled receptor affecting testis descent
- INSL:
-
insulin-like protein
- LGR:
-
leucine-rich repeat-containing G protein-coupled receptor
References
Le Roith D, Shiloach J, Roth J, Lesniak MA 1980 Evolutionary origins of vertebrate hormones: substances similar to mammalian insulins are native to unicellular eukaryotes. Proc Natl Acad Sci U S A 77: 6184–6188
Konopinska D, Rosinski G, Sobotka W 1992 Insect peptide hormones, an overview of the present literature. Int J Pept Protein Res 39: 1–11
Smit AB, van Marle A, van Elk R, Bogerd J, van Heerikhuizen H, Geraerts WP 1993 Evolutionary conservation of the insulin gene structure in invertebrates: cloning of the gene encoding molluscan insulin-related peptide III from Lymnaea stagnalis. J Mol Endocrinol 11: 103–113
Ishizaki H, Suzuki A 1994 The brain secretory peptides that control moulting and metamorphosis of the silkmoth, Bombyx mori. Int J Dev Bil 38: 301–310
Duret L, Guex N, Peitsch MC, Bairoch A 1998 New insulin-like proteins with atypical disulfide bond pattern characterized in Caenorhabditis elegans by comparative sequence analysis and homology modeling. Genome Res 8: 348–353
Reinecke M, Eppler E, David I, Georges D 1999 Immunohistochemical evidence for the presence, localization and partial coexistence of insulin, insulin-like growth factor I and relaxin in the protochordate Ciona intestinalis. Cell Tissue Res 295: 331–338
Blundell TL, Humbel RE 1980 Hormone families: pancreatic hormones and homologous growth factors. Nature 287: 781–787
Burkhardt E, Adham IM, Brosig B, Gastmann A, Mattei MG, Engel W 1994 Structural organization of the porcine and human genes coding for a Leydig cell-specific insulin-like peptide (LEY I-L) and chromosomal localization of the human gene (INSL3). Genomics 20: 13–19
Zimmermann S, Steding G, Emmen JM, Brinkmann AO, Nayernia K, Holstein AF, Engel W, Adham IM 1999 Targeted disruption of the Insl3 gene causes bilateral cryptorchidism. Mol Endocrinol 13: 681–691
Chassin D, Laurent A, Janneau JL, Berger R, Bellet D 1995 Cloning of a new member of the Insulin gene superfamily (INSL4) expressed in human placenta. Genomics 29: 465–470
Koman A, Cazaubon S, Couraud PO, Ullrich A, Strosberg AD 1996 Molecular characterization and in vitro biological activity of placentin, a new member of the insulin gene family. J Biol Chem 271: 20238–20241
Conklin D, Lofton-Day CE, Haldeman BA, Ching A, Whitmore TE, Lok S, Jaspers S 1999 Identification of INSL5, a new member of the insulin superfamily. Genomics 60: 50–56
Hsu SY 1999 Cloning of two novel mammalian paralogs of relaxin/insulin family proteins and their expression in testis and kidney. Mol Endocrinol 13: 2163–2174
Kasik J, Muglia L, Stephan DA, Menon RK 2000 Identification, chromosomal mapping, and partial characterization of mouse Insl6: a new member of the insulin family. Endocrinology 141: 458–461
Lok S, Johnston DS, Conklin D, Lofton-Day CE, Adams RL, Jelmberg AC, Whitmore TE, Schrader S, Griswold MD, Jaspers SR 2000 Identification of INSL6, a new member of the insulin family that is expressed in the testis of the human and rat. Biol Reprod 62: 1593–1599
Bathgate RA, Samuel CS, Burazin TC, Layfield S, Claasz AA, Reytomas IG, Dawson NF, Zhao C, Bond C, Summers RJ, Parry LJ, Wade JD, Tregear GW 2002 Human relaxin gene 3 (H3) and the equivalent mouse relaxin (M3) gene. Novel members of the relaxin peptide family. J Biol Chem 277: 1148–1157
Kasik JW, Lu C, Menon RK 2000 The expanding insulin family: structural, genomic, and functional considerations. Pediatr Diabetes 1: 169–177
Kumagai J, Hsu SY, Matsumi H, Roh JS, Fu P, Wade JD, Bathgate RA, Hsueh AJ 2002 INSL3/Leydig insulin-like peptide activates the LGR8 receptor important in testis descent. J Biol Chem 277: 31283–31286
Adham I, Burkhardt E, Benahmed M, Engel W 1993 Cloning of a cDNA for a novel insulin-like peptide of the testicular Leydig cells. J Biol Chem 268: 26668–26672
Zimmermann S, Schottler P, Engel W, Adham IM 1997 Mouse Leydig insulin-like (Ley I-L) gene: structure and expression during testis and ovary development. Mol Reprod Dev 47: 30–38
Tashima LS, Mazoujian G, Bryant-Greenwood GD 1994 Human relaxins in normal, benign and neoplastic breast tissue. J Mol Endocrinol 12: 351–364
Pusch W, Balvers M, Ivell R 1996 Molecular cloning and expression of the relaxin-like factor from the mouse testis. Endocrinology 137: 3009–3013
Bathgate R, Balvers M, Hunt N, Ivell R 1996 Relaxin-like factor gene is highly expressed in the bovine ovary of the cycle and pregnancy: sequence and messenger ribonucleic acid analysis. Biol Reprod 55: 1452–1457
Balvers M, Spiess AN, Domagalski R, Hunt N, Kilic E, Mukhopadhyay AK, Hanks E, Charlton HM, Ivell R 1998 Relaxin-like factor expression as a marker of differentiation in the mouse testis and ovary. Endocrinology 139: 2960–2970
Spiess AN, Balvers M, Tena-Sempere M, Huhtaniemi I, Parry L, Ivell R 1999 Structure and expression of the rat relaxin-like factor (RLF) gene. Mol Reprod Dev 54: 319–325
Nef S, Parada LF 1999 Cryptorchidism in mice mutant for Insl3. Nat Genet 22: 295–299
Overbeek PA, Gorlov IP, Sutherland RW, Houston JB, Harrison WR, Boettger-Tong HL, Bishop CE, Agoulnik AI 2001 A transgenic insertion causing cryptorchidism in mice. Genesis 30: 26–35
Tomboc M, Lee PA, Mitwally MF, Schneck FX, Bellinger M, Witchel SF 2000 Insulin-like 3/relaxin-like factor gene mutations are associated with cryptorchidism. J Clin Endocrinol Metab 85: 4013–4018
Marin P, Ferlin A, Moro E, Rossi A, Bartoloni L, Rossato M, Foresta C 2001 Novel insulin-like 3 (INSL3) gene mutation associated with human cryptorchidism. Am J Med Genet 103: 348–349
Canto P, Escudero, Soderlund D, Nishimura E, Carranza-Lira S, Gutierrez J, Nava A, Mendez JP 2003 A novel mutation of the insulin-like 3 gene in patients with cryptorchidism. J Hum Genet 48: 86–90
Feng S, Cortessis VK, Hwang A, Hardy B, Koh CJ, Bogatcheva NV, Agoulnik AI 2004 Mutation analysis of INSL3 and GREAT/LGR8 genes in familial cryptorchidism. Urology 64: 1032–1036
Kawamura K, Kumagai J, Sudo S, Chun SY, Pisarska M, Morita H, Toppari J, Fu P, Wade JD, Bathgate RA, Hsueh AJ 2004 Paracrine regulation of mammalian oocyte maturation and male germ cell survival. Proc Natl Acad Sci U S A 101: 7323–7328
Bieche I, Laurent A, Laurendeau I, Duret L, Giovangrandi Y, Frendo JL, Olivi M, Fausser JL, Evain-Brion D, Vidaud M 2003 Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol Reprod 68: 1422–1429
Veitia R, Laurent A, Quintana-Murci L, Ottolenghi C, Fellous M, Vidaud M, McElreavey K 1998 The INSL4 gene maps close to WI-5527 at 9p24.1–>p23.3 clustered with two relaxin genes and outside the critical region for the monosomy 9p syndrome. Cytogenet Cell Genet 81: 275–277
Laurent A, Rouillac C, Delezoide AL, Giovangrandi Y, Vekemans M, Bellet D, Abitbol M, Vidaud M 1998 Insulin-like 4 (INSL4) gene expression in human embryonic and trophoblastic tissues. Mol Reprod Dev 51: 123–129
Janneau JL, Maldonado-Estrada J, Tachdjian G, Miran I, Motte N, Saulnier P, Sabourin JC, Cote JF, Simon B, Frydman R, Chaouat G, Bellet D 2002 Transcriptional expression of genes involved in cell invasion and migration by normal and tumoral trophoblast cells. J Clin Endocrinol Metab 87: 5336–5339
Lu C, Walker WH, Weisz OA, Watkins SC, Witchel SF, Sperling MA, Menon RK 2003 Insulin-like peptide 6 (Insl6): cellular and sub-cellular localization and evidence for post-translational modifications. Proceedings of the 85th Annual Meeting of the Endocrine Society, Philadelphia, PA, p. 178
Hsu SY, Kudo M, Chen T, Nakabayashi K, Bhalla A, van der Spek PJ, van Duin M, Hsueh AJ 2000 The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol Endocrinol 14: 1257–1271
Hsu SY, Nakabayashi K, Nishi S, Kumagai J, Kudo M, Sherwood OD, Hsueh AJ 2002 Activation of orphan receptors by the hormone relaxin. Science 295: 671–674
Sherwood OD 2004 Relaxin's physiological roles and other diverse actions. Endocr Rev 25: 205–234
Hsu SY 2003 New insights into the evolution of the relaxin-LGR signaling system. Trend Endocrinol Metab 14: 303–309
Sudo S, Kumagai J, Nishi S, Layfield S, Ferraro T, Bathgate RA, Hsueh AJ 2003 H3 Relaxin is a specific ligand for LGR7 and activates the receptor by interacting with both the ectodomain and the exoloop 2. J Biol Chem 278: 7855–7862
Mazerbourg S, Bouley DM, Sudo S, Klein CA, Zhang JV, Kawamura K, Goodrich LV, Rayburn H, Tessier-Lavigne M, Hsueh AJ 2004 Leucine-rich repeat-containing, G protein-coupled receptor 4 null mice exhibit intrauterine growth retardation associated with embryonic and perinatal lethality. Mol Endocrinol 18: 2241–2254
Morita H, Mazerbourg S, Bouley DM, Luo CW, Kawamura K, Kuwabara Y, Baribault H, Tian H, Hsueh AJ 2004 Neonatal lethality of LGR5 null mice is associated with ankyloglossia and gastrointestinal distension. Mol Cell Biol 24: 9736–9743
Nef S, Verma-Kurvari S, Merenmies J, Vassalli JD, Efstratiadis A, Accili D, Parada LF 2003 Testis determination requires insulin receptor family function in mice. Nature 426: 291–295
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported in part by NIH-HD044436.
Rights and permissions
About this article
Cite this article
Lu, C., Lam, H. & Menon, R. New Members of the Insulin Family: Regulators of Metabolism, Growth and Now … Reproduction. Pediatr Res 57, 70–73 (2005). https://doi.org/10.1203/01.PDR.0000159573.55187.CA
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/01.PDR.0000159573.55187.CA
This article is cited by
-
Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4)
Nature Communications (2023)
-
Regulation of spermatogenesis and reproductive capacity by Igf3 in tilapia
Cellular and Molecular Life Sciences (2020)
-
Genomic analysis reveals selection in Chinese native black pig
Scientific Reports (2016)
-
Studies of the molecular mechanisms of action of relaxin on the adenylyl cyclase signaling system using synthetic peptides derived from the LGR7 relaxin receptor
Neuroscience and Behavioral Physiology (2007)